A multiple instance learning approach for sequence data with across bag dependencies
Paper and Code
Jan 30, 2016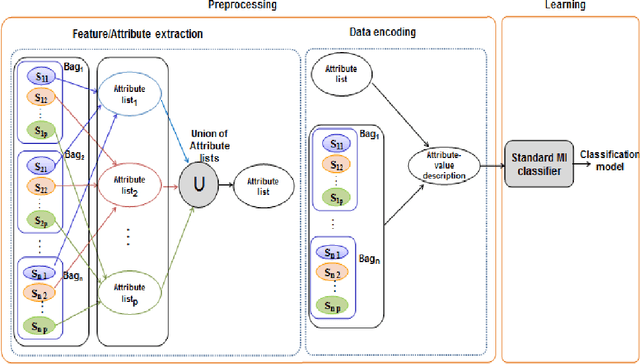
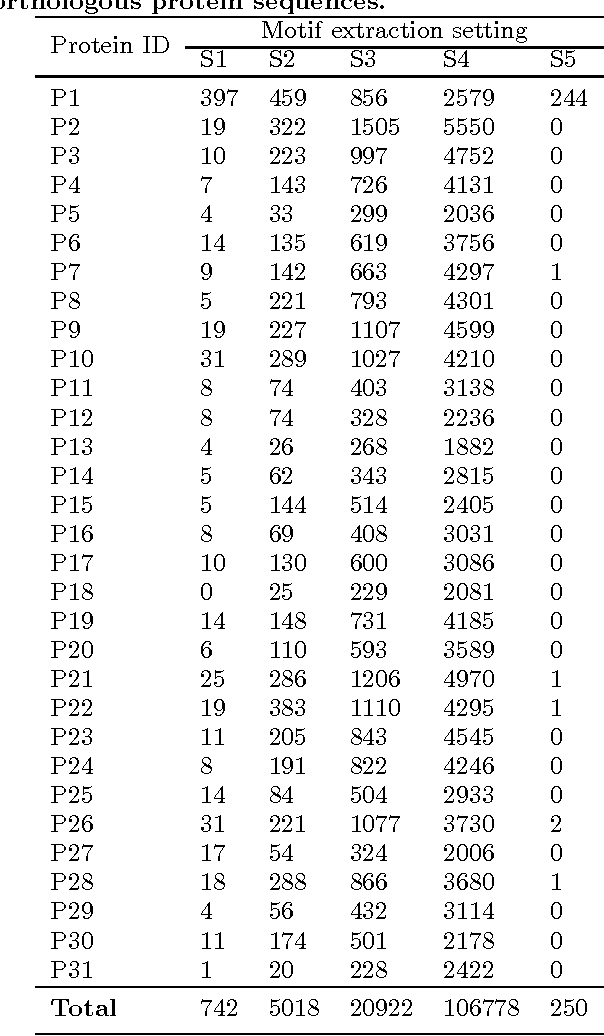
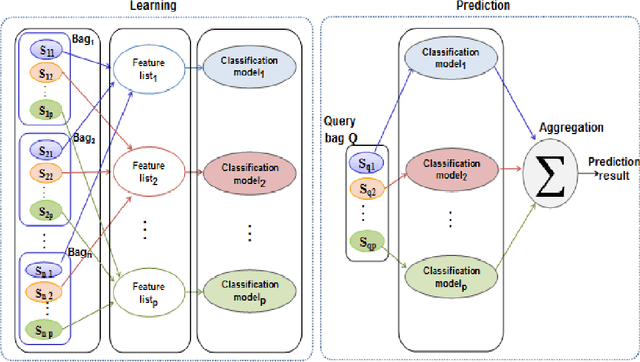
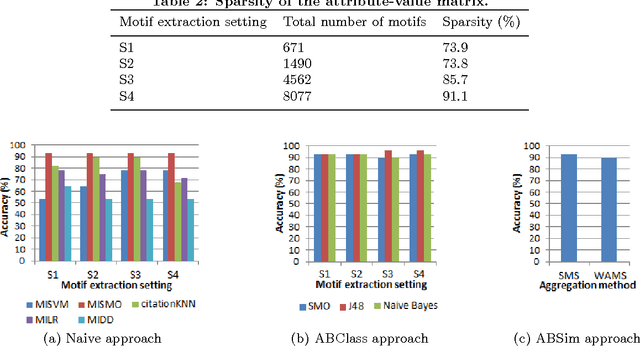
In Multiple Instance Learning (MIL) problem for sequence data, the learning data consist of a set of bags where each bag contains a set of instances/sequences. In many real world applications such as bioinformatics, web mining, and text mining, comparing a random couple of sequences makes no sense. In fact, each instance of each bag may have structural and/or temporal relation with other instances in other bags. Thus, the classification task should take into account the relation between semantically related instances across bags. In this paper, we present two novel MIL approaches for sequence data classification: (1) ABClass and (2) ABSim. In ABClass, each sequence is represented by one vector of attributes. For each sequence of the unknown bag, a discriminative classifier is applied in order to compute a partial classification result. Then, an aggregation method is applied to these partial results in order to generate the final result. In ABSim, we use a similarity measure between each sequence of the unknown bag and the corresponding sequences in the learning bags. An unknown bag is labeled with the bag that presents more similar sequences. We applied both approaches to the problem of bacterial Ionizing Radiation Resistance (IRR) prediction. We evaluated and discussed the proposed approaches on well known Ionizing Radiation Resistance Bacteria (IRRB) and Ionizing Radiation Sensitive Bacteria (IRSB) represented by primary structure of basal DNA repair proteins. The experimental results show that both ABClass and ABSim approaches are efficient.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge