A deep language model to predict metabolic network equilibria
Paper and Code
Dec 07, 2021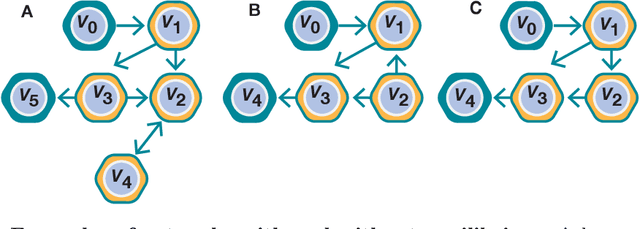
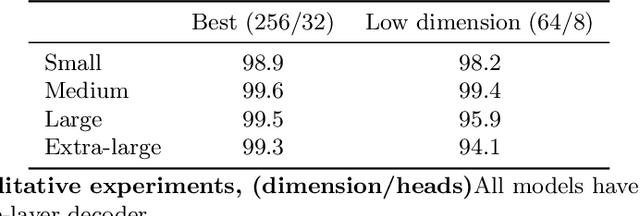


We show that deep learning models, and especially architectures like the Transformer, originally intended for natural language, can be trained on randomly generated datasets to predict to very high accuracy both the qualitative and quantitative features of metabolic networks. Using standard mathematical techniques, we create large sets (40 million elements) of random networks that can be used to train our models. These trained models can predict network equilibrium on random graphs in more than 99% of cases. They can also generalize to graphs with different structure than those encountered at training. Finally, they can predict almost perfectly the equilibria of a small set of known biological networks. Our approach is both very economical in experimental data and uses only small and shallow deep-learning model, far from the large architectures commonly used in machine translation. Such results pave the way for larger use of deep learning models for problems related to biological networks in key areas such as quantitative systems pharmacology, systems biology, and synthetic biology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge