3D Cell Nuclei Segmentation with Balanced Graph Partitioning
Paper and Code
Feb 17, 2017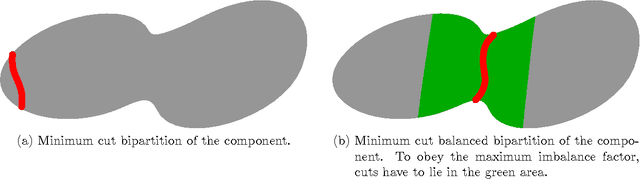
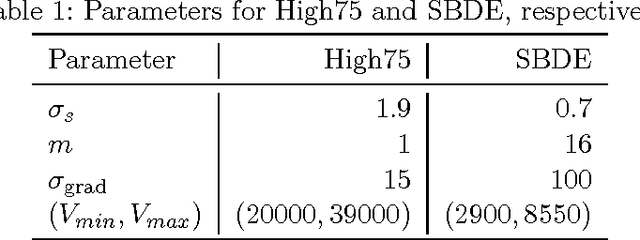
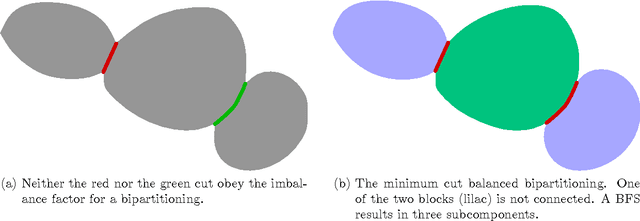

Cell nuclei segmentation is one of the most important tasks in the analysis of biomedical images. With ever-growing sizes and amounts of three-dimensional images to be processed, there is a need for better and faster segmentation methods. Graph-based image segmentation has seen a rise in popularity in recent years, but is seen as very costly with regard to computational demand. We propose a new segmentation algorithm which overcomes these limitations. Our method uses recursive balanced graph partitioning to segment foreground components of a fast and efficient binarization. We construct a model for the cell nuclei to guide the partitioning process. Our algorithm is compared to other state-of-the-art segmentation algorithms in an experimental evaluation on two sets of realistically simulated inputs. Our method is faster, has similar or better quality and an acceptable memory overhead.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge