Zhuowei Cheng
How Much Does It Hurt: A Deep Learning Framework for Chronic Pain Score Assessment
Sep 22, 2020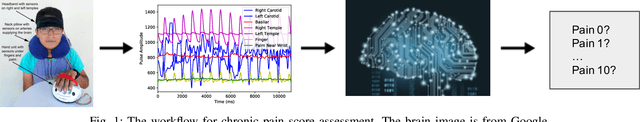
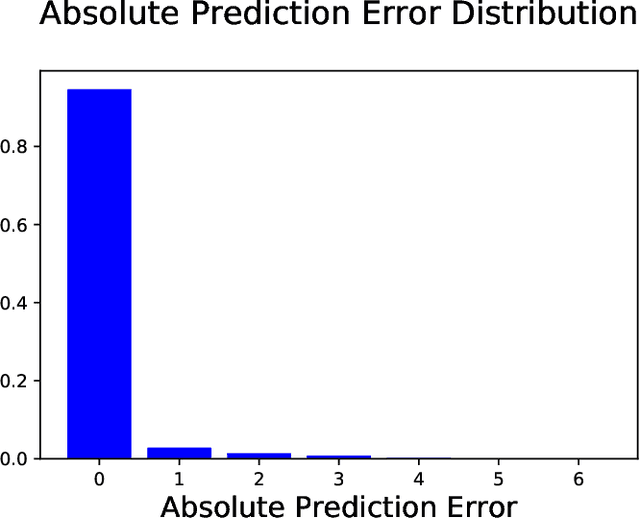

Abstract:Chronic pain is defined as pain that lasts or recurs for more than 3 to 6 months, often long after the injury or illness that initially caused the pain has healed. The "gold standard" for chronic pain assessment remains self report and clinical assessment via a biopsychosocial interview, since there has been no device that can measure it. A device to measure pain would be useful not only for clinical assessment, but potentially also as a biofeedback device leading to pain reduction. In this paper we propose an end-to-end deep learning framework for chronic pain score assessment. Our deep learning framework splits the long time-course data samples into shorter sequences, and uses Consensus Prediction to classify the results. We evaluate the performance of our framework on two chronic pain score datasets collected from two iterations of prototype Pain Meters that we have developed to help chronic pain subjects better understand their health condition.
A Deep Learning Framework for Classification of in vitro Multi-Electrode Array Recordings
Jun 05, 2019



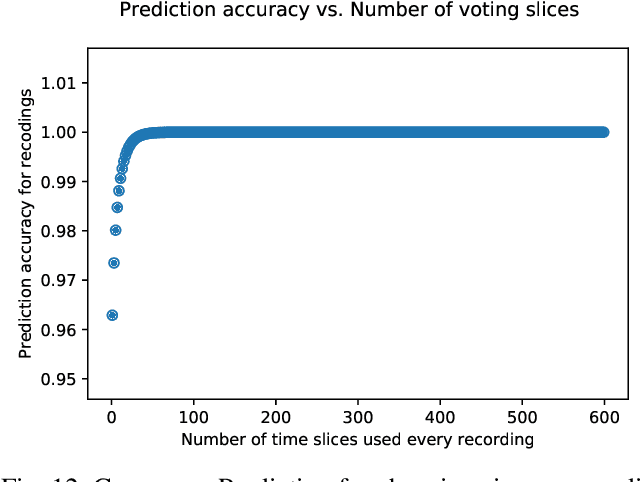
Abstract:Multi-Electrode Arrays (MEAs) have been widely used to record neuronal activities, which could be used in the diagnosis of gene defects and drug effects. In this paper, we address the problem of classifying in vitro MEA recordings of mouse and human neuronal cultures from different genotypes, where there is no easy way to directly utilize raw sequences as inputs to train an end-to-end classification model. While carefully extracting some features by hand could partially solve the problem, this approach suffers from obvious drawbacks such as difficulty of generalizing. We propose a deep learning framework to address this challenge. Our approach correctly classifies neuronal culture data prepared from two different genotypes -- a mouse Knockout of the delta-catenin gene and human induced Pluripotent Stem Cell-derived neurons from Williams syndrome. By splitting the long recordings into short slices for training, and applying Consensus Prediction during testing, our deep learning approach improves the prediction accuracy by 16.69% compared with feature based Logistic Regression for mouse MEA recordings. We further achieve an accuracy of 95.91% using Consensus Prediction in one subset of mouse MEA recording data, which were all recorded at six days in vitro. As high-density MEA recordings become more widely available, this approach could be generalized for classification of neurons carrying different mutations and classification of drug responses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge