Zaynaf Salaymang
Label-free segmentation from cardiac ultrasound using self-supervised learning
Oct 10, 2022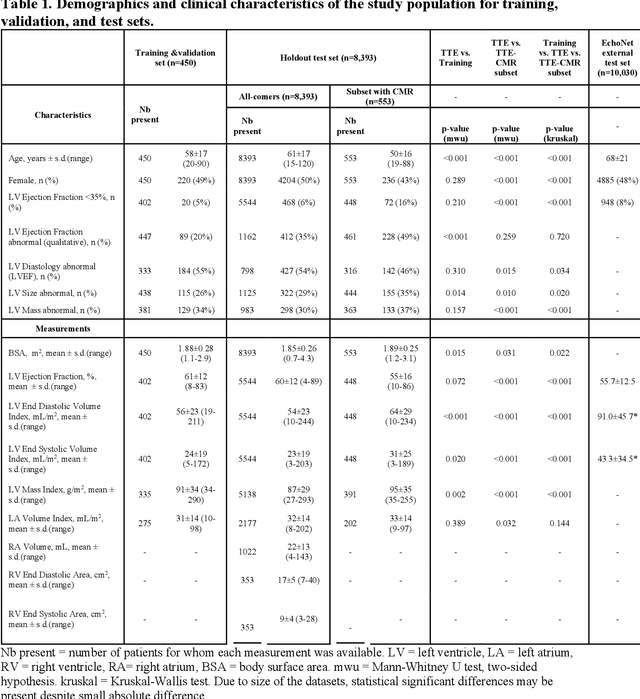
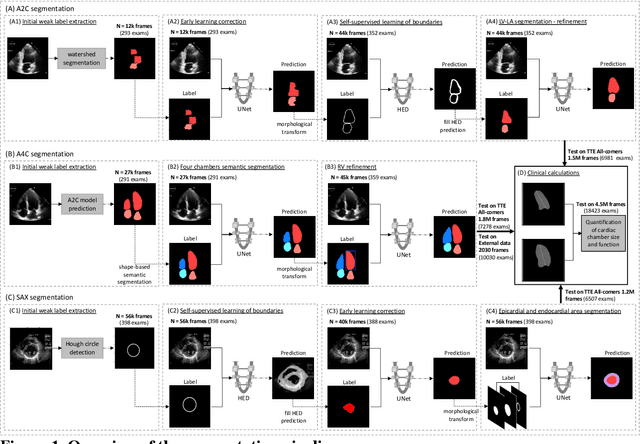
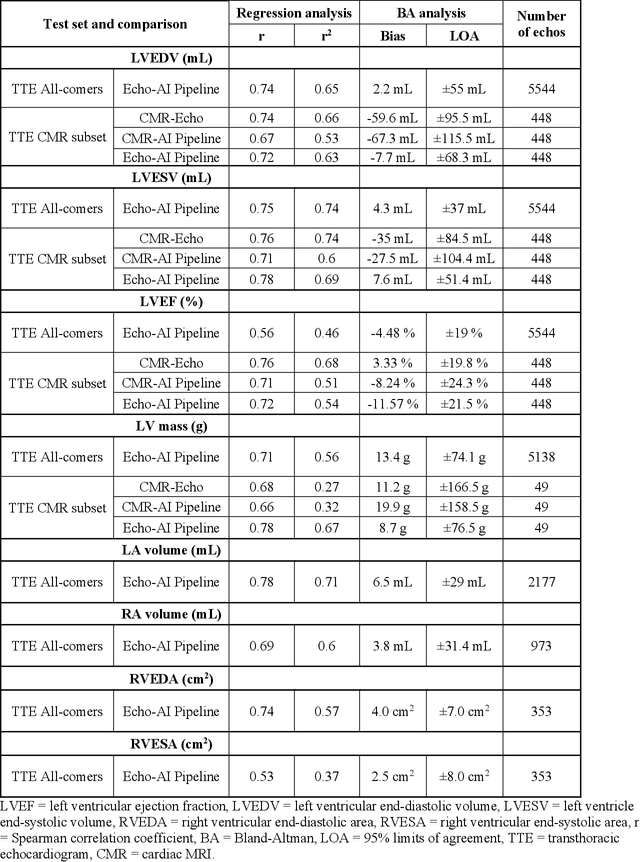
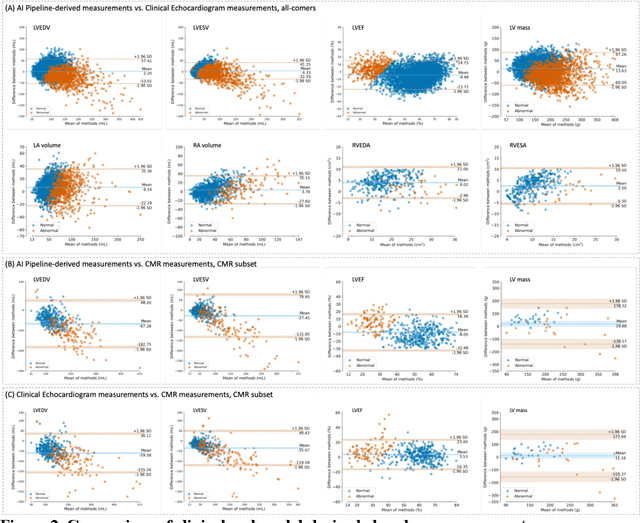
Abstract:Background: Segmentation and measurement of cardiac chambers is critical in echocardiography but is also laborious and poorly reproducible. Neural networks can assist, but supervised approaches require the same laborious manual annotations, while unsupervised approaches have fared poorly in ultrasound to date. Objectives: We built a pipeline for self-supervised (no manual labels required) segmentation of cardiac chambers, combining computer vision, clinical domain knowledge, and deep learning. Methods: We trained on 450 echocardiograms (145,000 images) and tested on 8,393 echocardiograms (4,476,266 images; mean age 61 years, 51% female), using the resulting segmentations to calculate structural and functional measurements. We also tested our pipeline against external images from an additional 10,030 patients (20,060 images) with available manual tracings of the left ventricle. Results: r2 between clinically measured and pipeline-predicted measurements were similar to reported inter-clinician variation for LVESV and LVEDV (pipeline vs. clinical r2= 0.74 and r2=0.65, respectively), LVEF and LV mass (r2= 0.46 and r2=0.54), left and right atrium volumes (r2=0.7 and r2=0.6), and right ventricle area (r2=0.47). When binarized into normal vs. abnormal categories, average accuracy was 0.81 (range 0.71-0.95). A subset of the test echocardiograms (n=553) had corresponding cardiac MRI; correlation between pipeline and CMR measurements was similar to that between clinical echocardiogram and CMR. Finally, in the external dataset, our pipeline accurately segments the left ventricle with an average Dice score of 0.83 (95% CI 0.83). Conclusions: Our results demonstrate a human-label-free, valid, and scalable method for segmentation from ultrasound, a noisy but globally important imaging modality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge