Zahed Rahmati
Drug-Target Interaction/Affinity Prediction: Deep Learning Models and Advances Review
Feb 21, 2025Abstract:Drug discovery remains a slow and expensive process that involves many steps, from detecting the target structure to obtaining approval from the Food and Drug Administration (FDA), and is often riddled with safety concerns. Accurate prediction of how drugs interact with their targets and the development of new drugs by using better methods and technologies have immense potential to speed up this process, ultimately leading to faster delivery of life-saving medications. Traditional methods used for drug-target interaction prediction show limitations, particularly in capturing complex relationships between drugs and their targets. As an outcome, deep learning models have been presented to overcome the challenges of interaction prediction through their precise and efficient end results. By outlining promising research avenues and models, each with a different solution but similar to the problem, this paper aims to give researchers a better idea of methods for even more accurate and efficient prediction of drug-target interaction, ultimately accelerating the development of more effective drugs. A total of 180 prediction methods for drug-target interactions were analyzed throughout the period spanning 2016 to 2025 using different frameworks based on machine learning, mainly deep learning and graph neural networks. Additionally, this paper discusses the novelty, architecture, and input representation of these models.
Edge2Node: Reducing Edge Prediction to Node Classification
Nov 13, 2023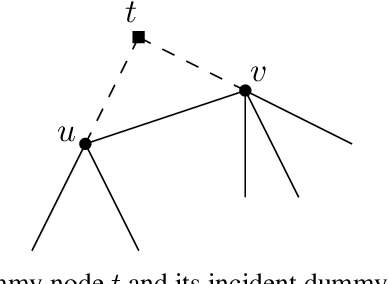
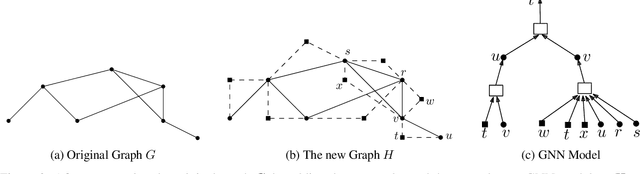
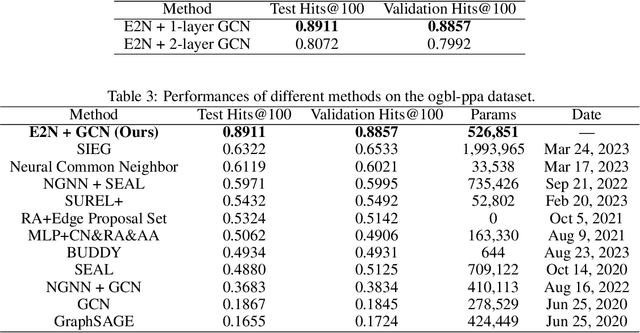
Abstract:Despite the success of graph neural network models in node classification, edge prediction (the task of predicting missing or potential links between nodes in a graph) remains a challenging problem for these models. A common approach for edge prediction is to first obtain the embeddings of two nodes, and then a predefined scoring function is used to predict the existence of an edge between the two nodes. In this paper, we introduce a new approach called Edge2Node (E2N) which directly obtains an embedding for each edge, without the need for a scoring function. To do this, we create a new graph H based on the graph G given for the edge prediction task, and then reduce the edge prediction task on G to a node classification task on H. Our E2N method can be easily applied to any edge prediction task with superior performance and lower computational costs. Our E2N method beats the best-known methods on the leaderboards for ogbl-ppa, ogbl-collab, and ogbl-ddi datasets by 25.89%, 24.19%, and 0.34% improvements, respectively.
BERT-DRE: BERT with Deep Recursive Encoder for Natural Language Sentence Matching
Nov 04, 2021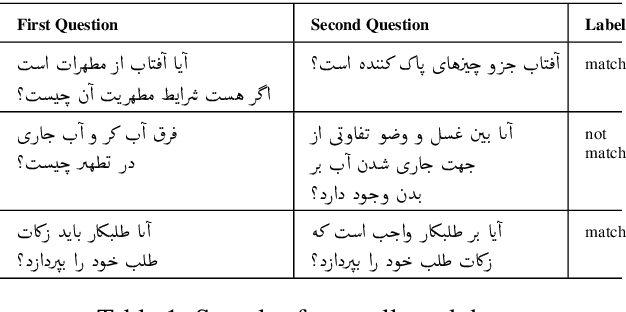
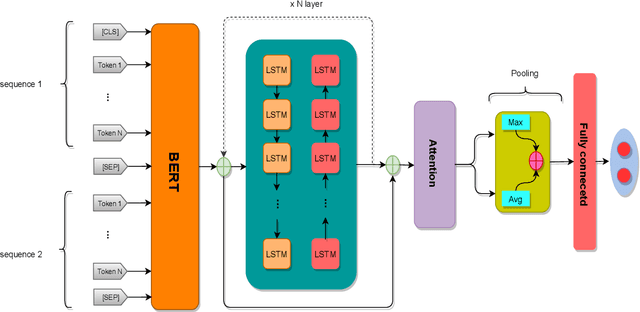
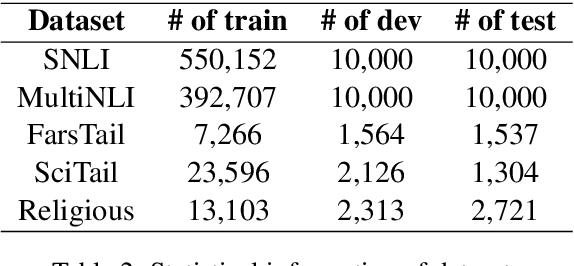
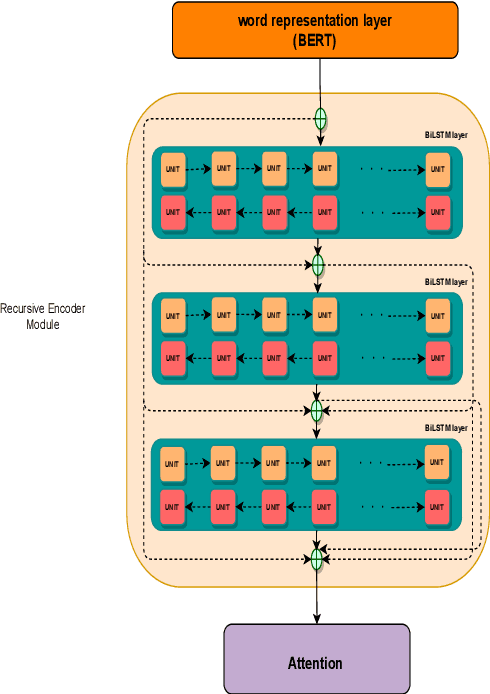
Abstract:This paper presents a deep neural architecture, for Natural Language Sentence Matching (NLSM) by adding a deep recursive encoder to BERT so called BERT with Deep Recursive Encoder (BERT-DRE). Our analysis of model behavior shows that BERT still does not capture the full complexity of text, so a deep recursive encoder is applied on top of BERT. Three Bi-LSTM layers with residual connection are used to design a recursive encoder and an attention module is used on top of this encoder. To obtain the final vector, a pooling layer consisting of average and maximum pooling is used. We experiment our model on four benchmarks, SNLI, FarsTail, MultiNLI, SciTail, and a novel Persian religious questions dataset. This paper focuses on improving the BERT results in the NLSM task. In this regard, comparisons between BERT-DRE and BERT are conducted, and it is shown that in all cases, BERT-DRE outperforms BERT. The BERT algorithm on the religious dataset achieved an accuracy of 89.70%, and BERT-DRE architectures improved to 90.29% using the same dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge