Yongjin Park
Causal Mediation Analysis Leveraging Multiple Types of Summary Statistics Data
Jan 24, 2019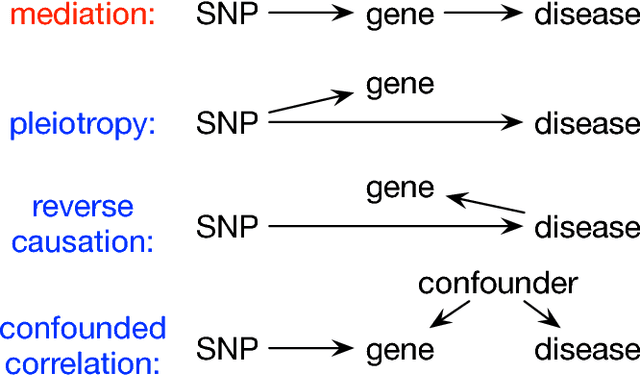
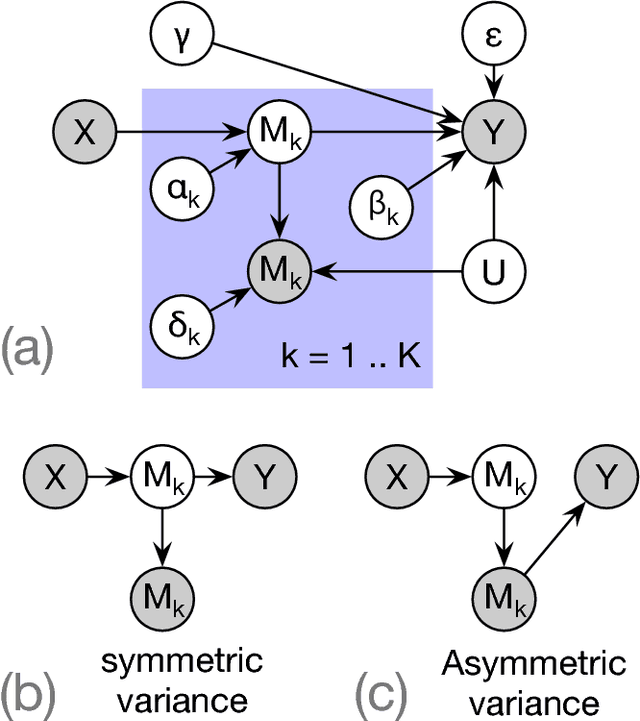
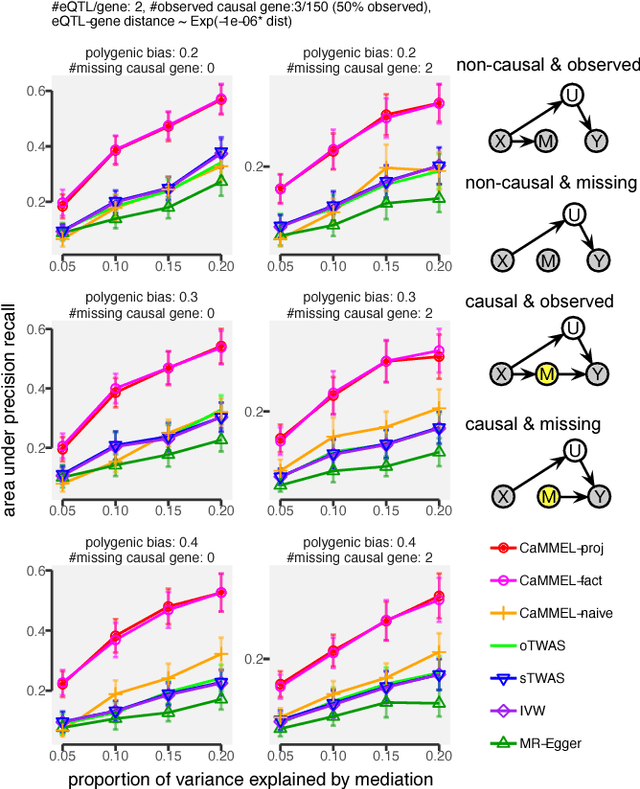
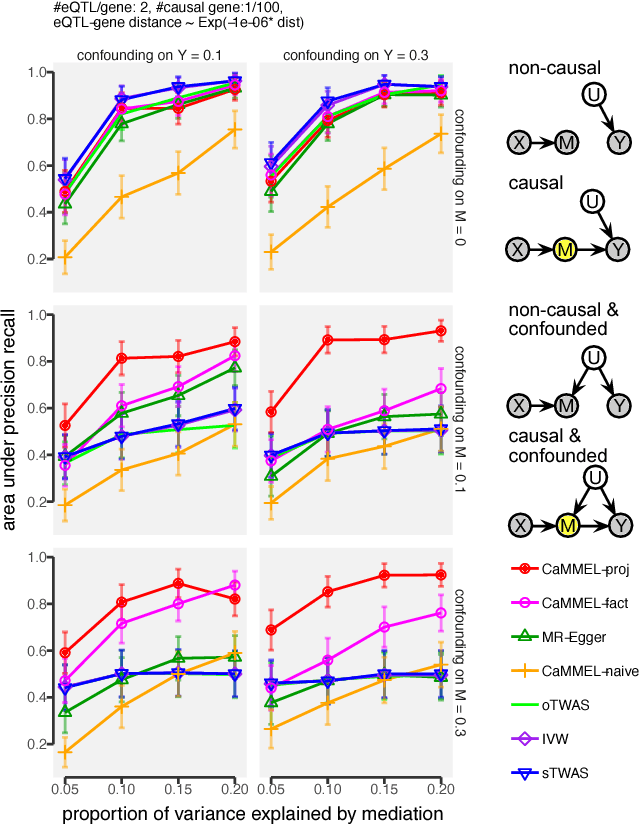
Abstract:Summary statistics of genome-wide association studies (GWAS) teach causal relationship between millions of genetic markers and tens and thousands of phenotypes. However, underlying biological mechanisms are yet to be elucidated. We can achieve necessary interpretation of GWAS in a causal mediation framework, looking to establish a sparse set of mediators between genetic and downstream variables, but there are several challenges. Unlike existing methods rely on strong and unrealistic assumptions, we tackle practical challenges within a principled summary-based causal inference framework. We analyzed the proposed methods in extensive simulations generated from real-world genetic data. We demonstrated only our approach can accurately redeem causal genes, even without knowing actual individual-level data, despite the presence of competing non-causal trails.
Fast and reliable inference algorithm for hierarchical stochastic block models
Nov 14, 2017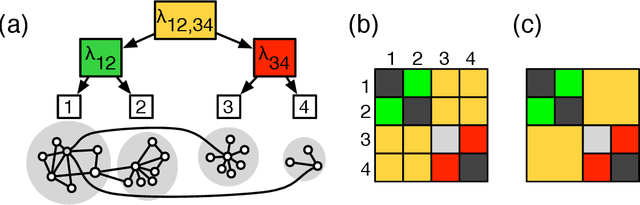
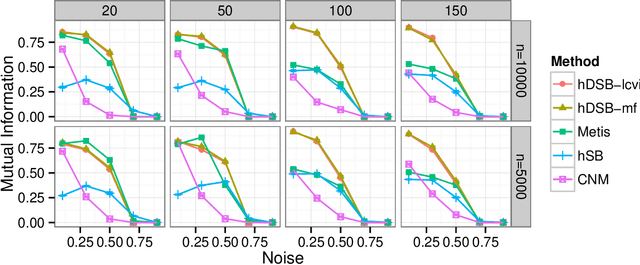
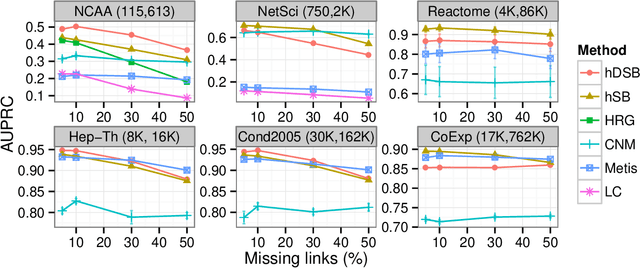
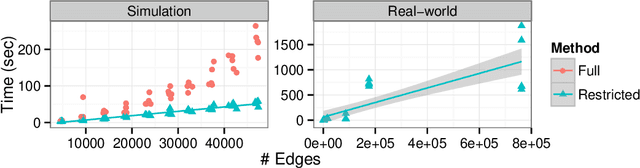
Abstract:Network clustering reveals the organization of a network or corresponding complex system with elements represented as vertices and interactions as edges in a (directed, weighted) graph. Although the notion of clustering can be somewhat loose, network clusters or groups are generally considered as nodes with enriched interactions and edges sharing common patterns. Statistical inference often treats groups as latent variables, with observed networks generated from latent group structure, termed a stochastic block model. Regardless of the definitions, statistical inference can be either translated to modularity maximization, which is provably an NP-complete problem. Here we present scalable and reliable algorithms that recover hierarchical stochastic block models fast and accurately. Our algorithm scales almost linearly in number of edges, and inferred models were more accurate that other scalable methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge