Yan-Li Lee
Data Skeleton Learning: Scalable Active Clustering with Sparse Graph Structures
Sep 10, 2025Abstract:In this work, we focus on the efficiency and scalability of pairwise constraint-based active clustering, crucial for processing large-scale data in applications such as data mining, knowledge annotation, and AI model pre-training. Our goals are threefold: (1) to reduce computational costs for iterative clustering updates; (2) to enhance the impact of user-provided constraints to minimize annotation requirements for precise clustering; and (3) to cut down memory usage in practical deployments. To achieve these aims, we propose a graph-based active clustering algorithm that utilizes two sparse graphs: one for representing relationships between data (our proposed data skeleton) and another for updating this data skeleton. These two graphs work in concert, enabling the refinement of connected subgraphs within the data skeleton to create nested clusters. Our empirical analysis confirms that the proposed algorithm consistently facilitates more accurate clustering with dramatically less input of user-provided constraints, and outperforms its counterparts in terms of computational performance and scalability, while maintaining robustness across various distance metrics.
Collaborative Filtering Approach to Link Prediction
Mar 14, 2021



Abstract:Link prediction is a fundamental challenge in network science. Among various methods, local similarity indices are widely used for their high cost-performance. However, the performance is less robust: for some networks local indices are highly competitive to state-of-the-art algorithms while for some other networks they are very poor. Inspired by techniques developed for recommender systems, we propose an enhancement framework for local indices based on collaborative filtering (CF). Considering the delicate but important difference between personalized recommendation and link prediction, we further propose an improved framework named as self-included collaborative filtering (SCF). The SCF framework significantly improved the accuracy and robustness of well-known local indices. The combination of SCF framework and a simple local index can produce an index with competitive performance and much lower complexity compared with elaborately-designed state-of-the-art algorithms.
A generalized method toward drug-target interaction prediction via low-rank matrix projection
May 03, 2018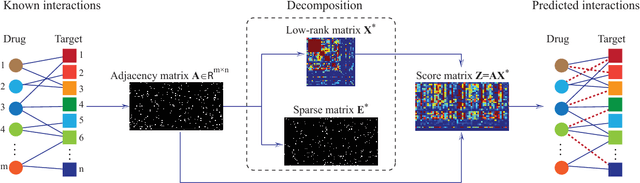
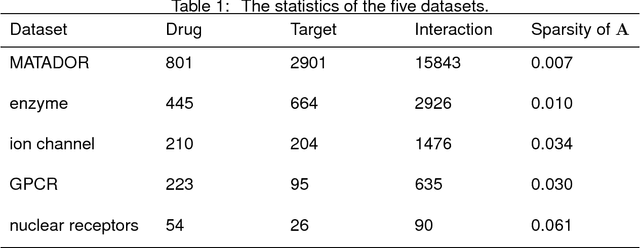
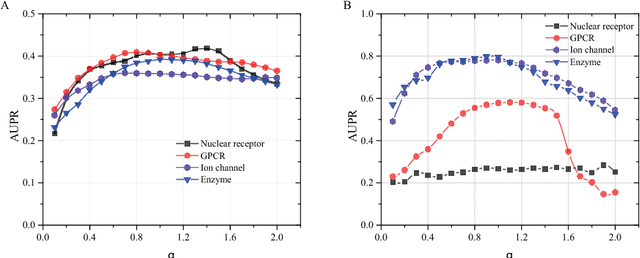
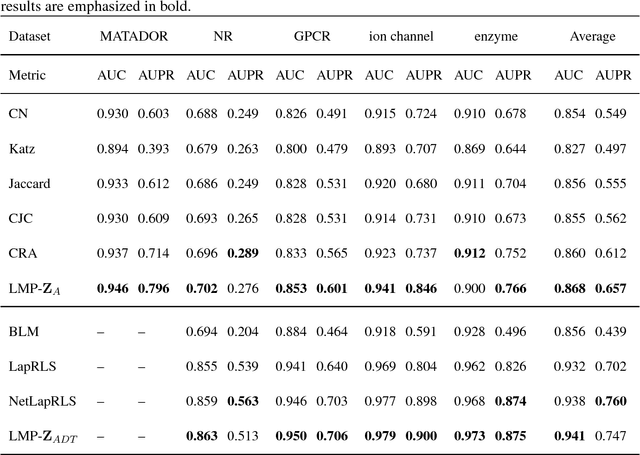
Abstract:Drug-target interaction (DTI) prediction plays a very important role in drug development and drug discovery. Biochemical experiments or \textit{in vitro} methods are very expensive, laborious and time-consuming. Therefore, \textit{in silico} approaches including docking simulation and machine learning have been proposed to solve this problem. In particular, machine learning approaches have attracted increasing attentions recently. However, in addition to the known drug-target interactions, most of the machine learning methods require extra characteristic information such as chemical structures, genome sequences, binding types and so on. Whenever such information is not available, they may perform poor. Very recently, the similarity-based link prediction methods were extended to bipartite networks, which can be applied to solve the DTI prediction problem by using topological information only. In this work, we propose a method based on low-rank matrix projection to solve the DTI prediction problem. On one hand, when there is no extra characteristic information of drugs or targets, the proposed method utilizes only the known interactions. On the other hand, the proposed method can also utilize the extra characteristic information when it is available and the performances will be remarkably improved. Moreover, the proposed method can predict the interactions associated with new drugs or targets of which we know nothing about their associated interactions, but only some characteristic information. We compare the proposed method with ten baseline methods, e.g., six similarity-based methods that utilize only the known interactions and four methods that utilize the extra characteristic information. The datasets and codes implementing the simulations are available at https://github.com/rathapech/DTI_LMP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge