Xiuzhuang Zhou
UniADC: A Unified Framework for Anomaly Detection and Classification
Nov 10, 2025Abstract:In this paper, we introduce the task of unified anomaly detection and classification, which aims to simultaneously detect anomalous regions in images and identify their specific categories. Existing methods typically treat anomaly detection and classification as separate tasks, thereby neglecting their inherent correlation, limiting information sharing, and resulting in suboptimal performance. To address this, we propose UniADC, a unified anomaly detection and classification model that can effectively perform both tasks with only a few or even no anomaly images. Specifically, UniADC consists of two key components: a training-free controllable inpainting network and a multi-task discriminator. The inpainting network can synthesize anomaly images of specific categories by repainting normal regions guided by anomaly priors, and can also repaint few-shot anomaly samples to augment the available anomaly data. The multi-task discriminator is then trained on these synthesized samples, enabling precise anomaly detection and classification by aligning fine-grained image features with anomaly-category embeddings. We conduct extensive experiments on three anomaly detection and classification datasets, including MVTec-FS, MTD, and WFDD, and the results demonstrate that UniADC consistently outperforms existing methods in anomaly detection, localization, and classification. The code is available at https://github.com/cnulab/UniADC.
JanusNet: Hierarchical Slice-Block Shuffle and Displacement for Semi-Supervised 3D Multi-Organ Segmentation
Aug 06, 2025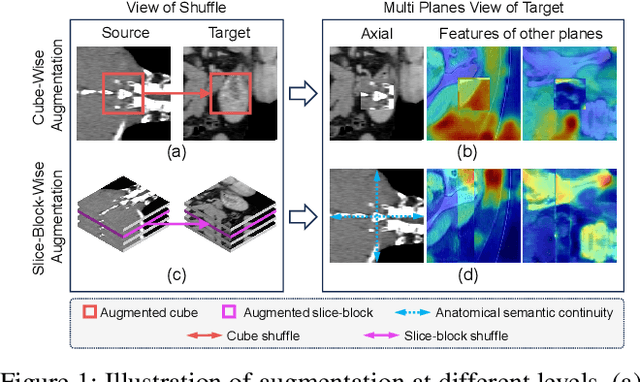
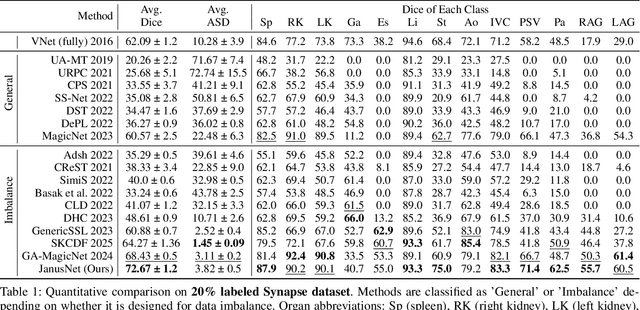
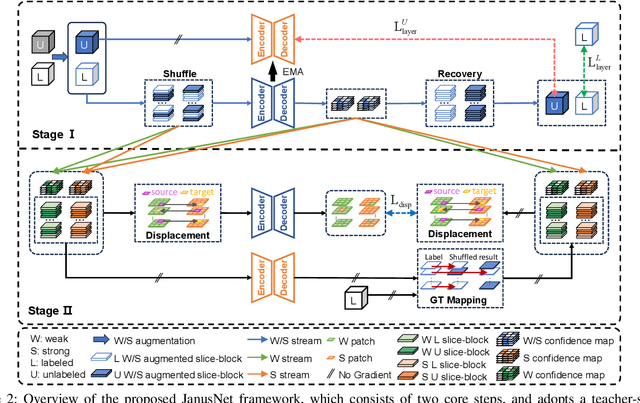
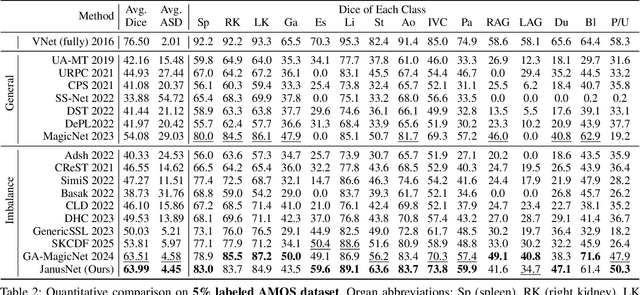
Abstract:Limited by the scarcity of training samples and annotations, weakly supervised medical image segmentation often employs data augmentation to increase data diversity, while randomly mixing volumetric blocks has demonstrated strong performance. However, this approach disrupts the inherent anatomical continuity of 3D medical images along orthogonal axes, leading to severe structural inconsistencies and insufficient training in challenging regions, such as small-sized organs, etc. To better comply with and utilize human anatomical information, we propose JanusNet}, a data augmentation framework for 3D medical data that globally models anatomical continuity while locally focusing on hard-to-segment regions. Specifically, our Slice-Block Shuffle step performs aligned shuffling of same-index slice blocks across volumes along a random axis, while preserving the anatomical context on planes perpendicular to the perturbation axis. Concurrently, the Confidence-Guided Displacement step uses prediction reliability to replace blocks within each slice, amplifying signals from difficult areas. This dual-stage, axis-aligned framework is plug-and-play, requiring minimal code changes for most teacher-student schemes. Extensive experiments on the Synapse and AMOS datasets demonstrate that JanusNet significantly surpasses state-of-the-art methods, achieving, for instance, a 4% DSC gain on the Synapse dataset with only 20% labeled data.
Fair Uncertainty Quantification for Depression Prediction
May 08, 2025Abstract:Trustworthy depression prediction based on deep learning, incorporating both predictive reliability and algorithmic fairness across diverse demographic groups, is crucial for clinical application. Recently, achieving reliable depression predictions through uncertainty quantification has attracted increasing attention. However, few studies have focused on the fairness of uncertainty quantification (UQ) in depression prediction. In this work, we investigate the algorithmic fairness of UQ, namely Equal Opportunity Coverage (EOC) fairness, and propose Fair Uncertainty Quantification (FUQ) for depression prediction. FUQ pursues reliable and fair depression predictions through group-based analysis. Specifically, we first group all the participants by different sensitive attributes and leverage conformal prediction to quantify uncertainty within each demographic group, which provides a theoretically guaranteed and valid way to quantify uncertainty for depression prediction and facilitates the investigation of fairness across different demographic groups. Furthermore, we propose a fairness-aware optimization strategy that formulates fairness as a constrained optimization problem under EOC constraints. This enables the model to preserve predictive reliability while adapting to the heterogeneous uncertainty levels across demographic groups, thereby achieving optimal fairness. Through extensive evaluations on several visual and audio depression datasets, our approach demonstrates its effectiveness.
$L^2$FMamba: Lightweight Light Field Image Super-Resolution with State Space Model
Mar 25, 2025Abstract:Transformers bring significantly improved performance to the light field image super-resolution task due to their long-range dependency modeling capability. However, the inherently high computational complexity of their core self-attention mechanism has increasingly hindered their advancement in this task. To address this issue, we first introduce the LF-VSSM block, a novel module inspired by progressive feature extraction, to efficiently capture critical long-range spatial-angular dependencies in light field images. LF-VSSM successively extracts spatial features within sub-aperture images, spatial-angular features between sub-aperture images, and spatial-angular features between light field image pixels. On this basis, we propose a lightweight network, $L^2$FMamba (Lightweight Light Field Mamba), which integrates the LF-VSSM block to leverage light field features for super-resolution tasks while overcoming the computational challenges of Transformer-based approaches. Extensive experiments on multiple light field datasets demonstrate that our method reduces the number of parameters and complexity while achieving superior super-resolution performance with faster inference speed.
Billet Number Recognition Based on Test-Time Adaptation
Feb 13, 2025Abstract:During the steel billet production process, it is essential to recognize machine-printed or manually written billet numbers on moving billets in real-time. To address the issue of low recognition accuracy for existing scene text recognition methods, caused by factors such as image distortions and distribution differences between training and test data, we propose a billet number recognition method that integrates test-time adaptation with prior knowledge. First, we introduce a test-time adaptation method into a model that uses the DB network for text detection and the SVTR network for text recognition. By minimizing the model's entropy during the testing phase, the model can adapt to the distribution of test data without the need for supervised fine-tuning. Second, we leverage the billet number encoding rules as prior knowledge to assess the validity of each recognition result. Invalid results, which do not comply with the encoding rules, are replaced. Finally, we introduce a validation mechanism into the CTC algorithm using prior knowledge to address its limitations in recognizing damaged characters. Experimental results on real datasets, including both machine-printed billet numbers and handwritten billet numbers, show significant improvements in evaluation metrics, validating the effectiveness of the proposed method.
Conformal Depression Prediction
May 29, 2024



Abstract:While existing depression recognition methods based on deep learning show promise, their practical application is hindered by the lack of trustworthiness, as these deep models are often deployed as \textit{black box} models, leaving us uncertain about the confidence of the model predictions. For high-risk clinical applications like depression recognition, uncertainty quantification is essential in decision-making. In this paper, we introduce conformal depression prediction (CDP), a depression recognition method with uncertainty quantification based on conformal prediction (CP), giving valid confidence intervals with theoretical coverage guarantees for the model predictions. CDP is a plug-and-play module that requires neither model retraining nor an assumption about the depression data distribution. As CDP provides only an average performance guarantee across all inputs rather than per-input performance guarantee, we propose CDP-ACC, an improved conformal prediction with approximate conditional coverage. CDP-ACC firstly estimates the prediction distribution through neighborhood relaxation, and then introduces a conformal score function by constructing nested sequences, so as to provide tighter prediction interval for each specific input. We empirically demonstrate the application of uncertainty quantification in depression recognition, and the effectiveness and superiority of CDP and CDP-ACC on the AVEC 2013 and AVEC 2014 datasets
MediCLIP: Adapting CLIP for Few-shot Medical Image Anomaly Detection
May 18, 2024



Abstract:In the field of medical decision-making, precise anomaly detection in medical imaging plays a pivotal role in aiding clinicians. However, previous work is reliant on large-scale datasets for training anomaly detection models, which increases the development cost. This paper first focuses on the task of medical image anomaly detection in the few-shot setting, which is critically significant for the medical field where data collection and annotation are both very expensive. We propose an innovative approach, MediCLIP, which adapts the CLIP model to few-shot medical image anomaly detection through self-supervised fine-tuning. Although CLIP, as a vision-language model, demonstrates outstanding zero-/fewshot performance on various downstream tasks, it still falls short in the anomaly detection of medical images. To address this, we design a series of medical image anomaly synthesis tasks to simulate common disease patterns in medical imaging, transferring the powerful generalization capabilities of CLIP to the task of medical image anomaly detection. When only few-shot normal medical images are provided, MediCLIP achieves state-of-the-art performance in anomaly detection and location compared to other methods. Extensive experiments on three distinct medical anomaly detection tasks have demonstrated the superiority of our approach. The code is available at https://github.com/cnulab/MediCLIP.
RealNet: A Feature Selection Network with Realistic Synthetic Anomaly for Anomaly Detection
Mar 09, 2024Abstract:Self-supervised feature reconstruction methods have shown promising advances in industrial image anomaly detection and localization. Despite this progress, these methods still face challenges in synthesizing realistic and diverse anomaly samples, as well as addressing the feature redundancy and pre-training bias of pre-trained feature. In this work, we introduce RealNet, a feature reconstruction network with realistic synthetic anomaly and adaptive feature selection. It is incorporated with three key innovations: First, we propose Strength-controllable Diffusion Anomaly Synthesis (SDAS), a diffusion process-based synthesis strategy capable of generating samples with varying anomaly strengths that mimic the distribution of real anomalous samples. Second, we develop Anomaly-aware Features Selection (AFS), a method for selecting representative and discriminative pre-trained feature subsets to improve anomaly detection performance while controlling computational costs. Third, we introduce Reconstruction Residuals Selection (RRS), a strategy that adaptively selects discriminative residuals for comprehensive identification of anomalous regions across multiple levels of granularity. We assess RealNet on four benchmark datasets, and our results demonstrate significant improvements in both Image AUROC and Pixel AUROC compared to the current state-o-the-art methods. The code, data, and models are available at https://github.com/cnulab/RealNet.
Masked Contrastive Reconstruction for Cross-modal Medical Image-Report Retrieval
Dec 27, 2023Abstract:Cross-modal medical image-report retrieval task plays a significant role in clinical diagnosis and various medical generative tasks. Eliminating heterogeneity between different modalities to enhance semantic consistency is the key challenge of this task. The current Vision-Language Pretraining (VLP) models, with cross-modal contrastive learning and masked reconstruction as joint training tasks, can effectively enhance the performance of cross-modal retrieval. This framework typically employs dual-stream inputs, using unmasked data for cross-modal contrastive learning and masked data for reconstruction. However, due to task competition and information interference caused by significant differences between the inputs of the two proxy tasks, the effectiveness of representation learning for intra-modal and cross-modal features is limited. In this paper, we propose an efficient VLP framework named Masked Contrastive and Reconstruction (MCR), which takes masked data as the sole input for both tasks. This enhances task connections, reducing information interference and competition between them, while also substantially decreasing the required GPU memory and training time. Moreover, we introduce a new modality alignment strategy named Mapping before Aggregation (MbA). Unlike previous methods, MbA maps different modalities to a common feature space before conducting local feature aggregation, thereby reducing the loss of fine-grained semantic information necessary for improved modality alignment. Qualitative and quantitative experiments conducted on the MIMIC-CXR dataset validate the effectiveness of our approach, demonstrating state-of-the-art performance in medical cross-modal retrieval tasks.
Weakly Supervised Anomaly Detection for Chest X-Ray Image
Nov 18, 2023Abstract:Chest X-Ray (CXR) examination is a common method for assessing thoracic diseases in clinical applications. While recent advances in deep learning have enhanced the significance of visual analysis for CXR anomaly detection, current methods often miss key cues in anomaly images crucial for identifying disease regions, as they predominantly rely on unsupervised training with normal images. This letter focuses on a more practical setup in which few-shot anomaly images with only image-level labels are available during training. For this purpose, we propose WSCXR, a weakly supervised anomaly detection framework for CXR. WSCXR firstly constructs sets of normal and anomaly image features respectively. It then refines the anomaly image features by eliminating normal region features through anomaly feature mining, thus fully leveraging the scarce yet crucial features of diseased areas. Additionally, WSCXR employs a linear mixing strategy to augment the anomaly features, facilitating the training of anomaly detector with few-shot anomaly images. Experiments on two CXR datasets demonstrate the effectiveness of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge