William E. Carson IV
Multiple Domain Causal Networks
May 13, 2022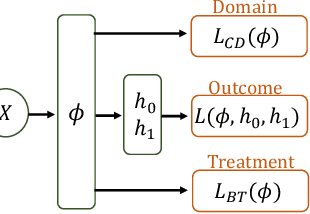


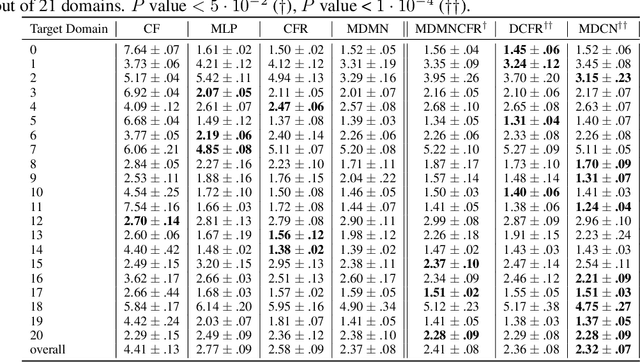
Abstract:Observational studies are regarded as economic alternatives to randomized trials, often used in their stead to investigate and determine treatment efficacy. Due to lack of sample size, observational studies commonly combine data from multiple sources or different sites/centers. Despite the benefits of an increased sample size, a naive combination of multicenter data may result in incongruities stemming from center-specific protocols for generating cohorts or reactions towards treatments distinct to a given center, among other things. These issues arise in a variety of other contexts, including capturing a treatment effect related to an individual's unique biological characteristics. Existing methods for estimating heterogeneous treatment effects have not adequately addressed the multicenter context, but rather treat it simply as a means to obtain sufficient sample size. Additionally, previous approaches to estimating treatment effects do not straightforwardly generalize to the multicenter design, especially when required to provide treatment insights for patients from a new, unobserved center. To address these shortcomings, we propose Multiple Domain Causal Networks (MDCN), an approach that simultaneously strengthens the information sharing between similar centers while addressing the selection bias in treatment assignment through learning of a new feature embedding. In empirical evaluations, MDCN is consistently more accurate when estimating the heterogeneous treatment effect in new centers compared to benchmarks that adjust solely based on treatment imbalance or general center differences. Finally, we justify our approach by providing theoretical analyses that demonstrate that MDCN improves on the generalization bound of the new, unobserved target center.
AugmentedPCA: A Python Package of Supervised and Adversarial Linear Factor Models
Jan 07, 2022



Abstract:Deep autoencoders are often extended with a supervised or adversarial loss to learn latent representations with desirable properties, such as greater predictivity of labels and outcomes or fairness with respects to a sensitive variable. Despite the ubiquity of supervised and adversarial deep latent factor models, these methods should demonstrate improvement over simpler linear approaches to be preferred in practice. This necessitates a reproducible linear analog that still adheres to an augmenting supervised or adversarial objective. We address this methodological gap by presenting methods that augment the principal component analysis (PCA) objective with either a supervised or an adversarial objective and provide analytic and reproducible solutions. We implement these methods in an open-source Python package, AugmentedPCA, that can produce excellent real-world baselines. We demonstrate the utility of these factor models on an open-source, RNA-seq cancer gene expression dataset, showing that augmenting with a supervised objective results in improved downstream classification performance, produces principal components with greater class fidelity, and facilitates identification of genes aligned with the principal axes of data variance with implications to development of specific types of cancer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge