Weihuang Xu
PRMI: A Dataset of Minirhizotron Images for Diverse Plant Root Study
Jan 20, 2022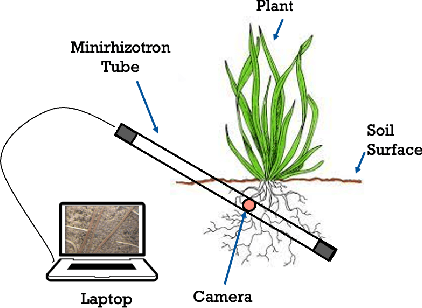
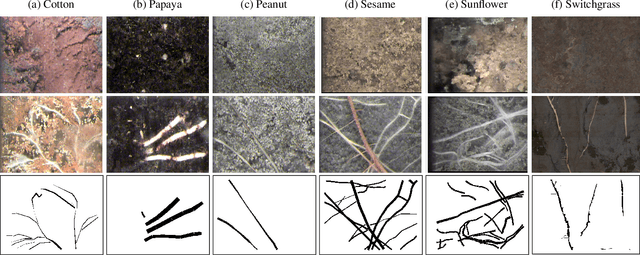
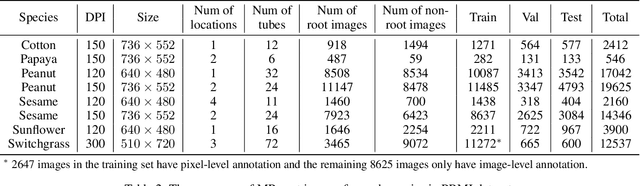
Abstract:Understanding a plant's root system architecture (RSA) is crucial for a variety of plant science problem domains including sustainability and climate adaptation. Minirhizotron (MR) technology is a widely-used approach for phenotyping RSA non-destructively by capturing root imagery over time. Precisely segmenting roots from the soil in MR imagery is a critical step in studying RSA features. In this paper, we introduce a large-scale dataset of plant root images captured by MR technology. In total, there are over 72K RGB root images across six different species including cotton, papaya, peanut, sesame, sunflower, and switchgrass in the dataset. The images span a variety of conditions including varied root age, root structures, soil types, and depths under the soil surface. All of the images have been annotated with weak image-level labels indicating whether each image contains roots or not. The image-level labels can be used to support weakly supervised learning in plant root segmentation tasks. In addition, 63K images have been manually annotated to generate pixel-level binary masks indicating whether each pixel corresponds to root or not. These pixel-level binary masks can be used as ground truth for supervised learning in semantic segmentation tasks. By introducing this dataset, we aim to facilitate the automatic segmentation of roots and the research of RSA with deep learning and other image analysis algorithms.
Weakly Supervised Minirhizotron Image Segmentation with MIL-CAM
Jul 30, 2020



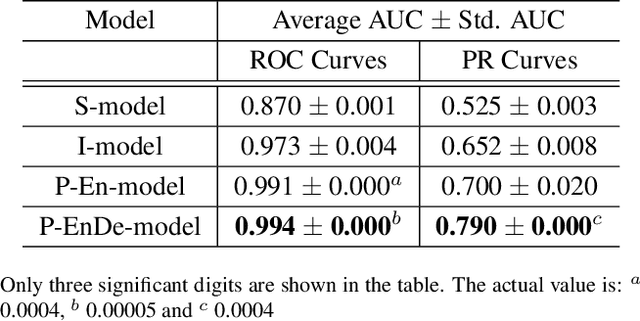
Abstract:We present a multiple instance learning class activation map (MIL-CAM) approach for pixel-level minirhizotron image segmentation given weak image-level labels. Minirhizotrons are used to image plant roots in situ. Minirhizotron imagery is often composed of soil containing a few long and thin root objects of small diameter. The roots prove to be challenging for existing semantic image segmentation methods to discriminate. In addition to learning from weak labels, our proposed MIL-CAM approach re-weights the root versus soil pixels during analysis for improved performance due to the heavy imbalance between soil and root pixels. The proposed approach outperforms other attention map and multiple instance learning methods for localization of root objects in minirhizotron imagery.
Histogram Layers for Texture Analysis
Jan 16, 2020



Abstract:We present a histogram layer for artificial neural networks (ANNs). An essential aspect of texture analysis is the extraction of features that describe the distribution of values in local spatial regions. The proposed histogram layer leverages the spatial distribution of features for texture analysis and parameters for the layer are estimated during backpropagation. We compare our method with state-of-the-art texture encoding methods such as the Deep Encoding Network (DEP) and Deep Texture Encoding Network (DeepTEN) on three texture datasets: (1) the Describable Texture Dataset (DTD); (2) an extension of the ground terrain in outdoor scenes (GTOS-mobile); (3) and a subset of the Materials in Context (MINC-2500) dataset. Results indicate that the inclusion of the proposed histogram layer improves performance. The source code for the histogram layer is publicly available.
Overcoming Small Minirhizotron Datasets Using Transfer Learning
Mar 22, 2019
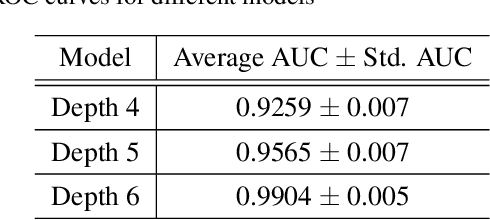

Abstract:Minirhizotron technology is widely used for studying the development of roots. Such systems collect visible-wavelength color imagery of plant roots in-situ by scanning an imaging system within a clear tube driven into the soil. Automated analysis of root systems could facilitate new scientific discoveries that would be critical to address the world's pressing food, resource, and climate issues. A key component of automated analysis of plant roots from imagery is the automated pixel-level segmentation of roots from their surrounding soil. Supervised learning techniques appear to be an appropriate tool for the challenge due to varying local soil and root conditions, however, lack of enough annotated training data is a major limitation due to the error-prone and time-consuming manually labeling process. In this paper, we investigate the use of deep neural networks based on the U-net architecture for automated, precise pixel-wise root segmentation in minirhizotron imagery. We compiled two minirhizotron image datasets to accomplish this study: one with 17,550 peanut root images and another with 28 switchgrass root images. Both datasets were paired with manually labeled ground truth masks. We trained three neural networks with different architectures on the larger peanut root dataset to explore the effect of the neural network depth on segmentation performance. To tackle the more limited switchgrass root dataset, we showed that models initialized with features pre-trained on the peanut dataset and then fine-tuned on the switchgrass dataset can improve segmentation performance significantly. We obtained 99\% segmentation accuracy in switchgrass imagery using only 21 training images. We also observed that features pre-trained on a closely related but relatively moderate size dataset like our peanut dataset are more effective than features pre-trained on the large but unrelated ImageNet dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge