Wee-Hong Ong
A Novel Skeleton-Based Human Activity Discovery Technique Using Particle Swarm Optimization with Gaussian Mutation
Jan 14, 2022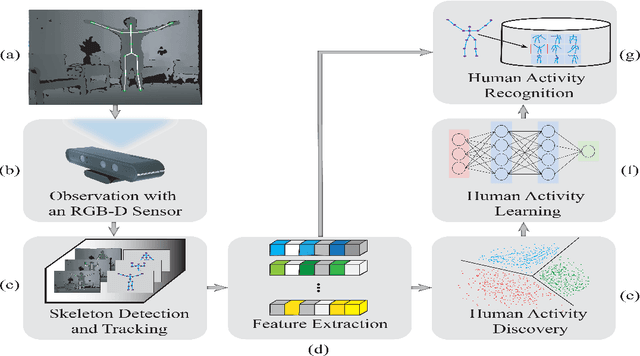
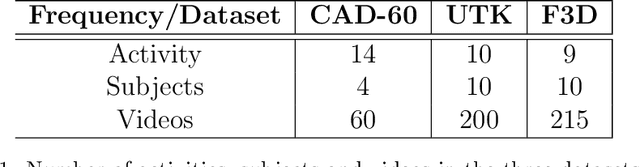
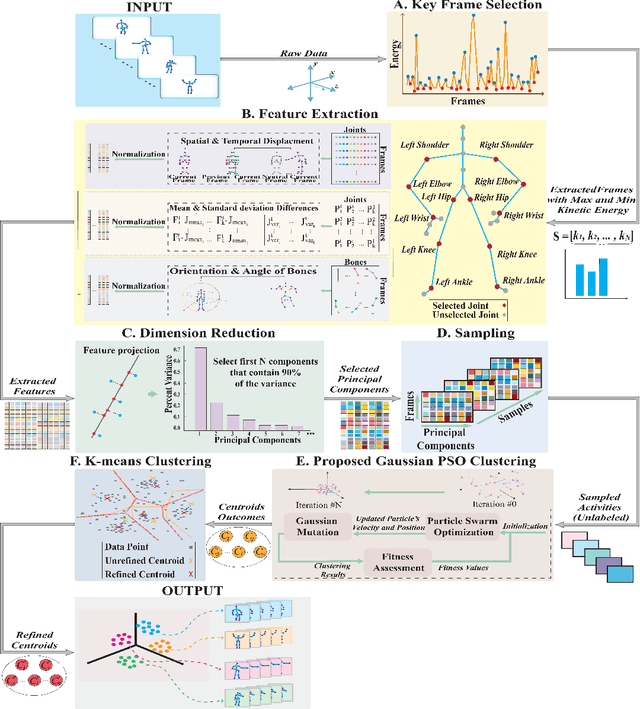
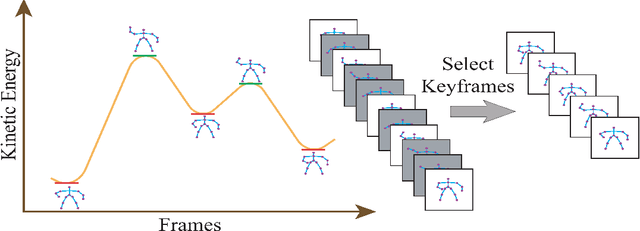
Abstract:Human activity discovery aims to distinguish the activities performed by humans, without any prior information of what defines each activity. Most methods presented in human activity recognition are supervised, where there are labeled inputs to train the system. In reality, it is difficult to label data because of its huge volume and the variety of activities performed by humans. In this paper, a novel unsupervised approach is proposed to perform human activity discovery in 3D skeleton sequences. First, important frames are selected based on kinetic energy. Next, the displacement of joints, set of statistical, angles, and orientation features are extracted to represent the activities information. Since not all extracted features have useful information, the dimension of features is reduced using PCA. Most human activity discovery proposed are not fully unsupervised. They use pre-segmented videos before categorizing activities. To deal with this, we used the fragmented sliding time window method to segment the time series of activities with some overlapping. Then, activities are discovered by a novel hybrid particle swarm optimization with a Gaussian mutation algorithm to avoid getting stuck in the local optimum. Finally, k-means is applied to the outcome centroids to overcome the slow rate of PSO. Experiments on three datasets have been presented and the results show the proposed method has superior performance in discovering activities in all evaluation parameters compared to the other state-of-the-art methods and has increased accuracy of at least 4 % on average. The code is available here: https://github.com/parhamhadikhani/Human-Activity-Discovery-HPGMK
Application of Computer Vision and Machine Learning for Digitized Herbarium Specimens: A Systematic Literature Review
Apr 18, 2021


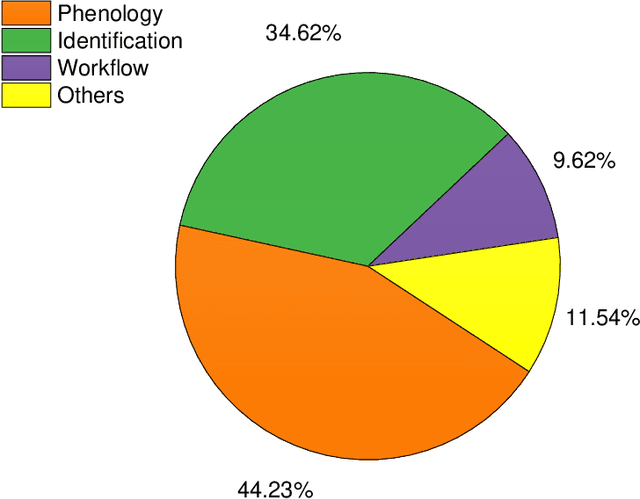
Abstract:Herbarium contains treasures of millions of specimens which have been preserved for several years for scientific studies. To speed up more scientific discoveries, a digitization of these specimens is currently on going to facilitate easy access and sharing of its data to a wider scientific community. Online digital repositories such as IDigBio and GBIF have already accumulated millions of specimen images yet to be explored. This presents a perfect time to automate and speed up more novel discoveries using machine learning and computer vision. In this study, a thorough analysis and comparison of more than 50 peer-reviewed studies which focus on application of computer vision and machine learning techniques to digitized herbarium specimen have been examined. The study categorizes different techniques and applications which have been commonly used and it also highlights existing challenges together with their possible solutions. It is our hope that the outcome of this study will serve as a strong foundation for beginners of the relevant field and will also shed more light for both computer science and ecology experts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge