Vladimír Boža
MACKO: Sparse Matrix-Vector Multiplication for Low Sparsity
Nov 17, 2025Abstract:Sparse Matrix-Vector Multiplication (SpMV) is a fundamental operation in the inference of sparse Large Language Models (LLMs). Because existing SpMV methods perform poorly under the low and unstructured sparsity (30-90%) commonly observed in pruned LLMs, unstructured pruning provided only limited memory reduction and speedup. We propose MACKO-SpMV, a GPU-optimized format and kernel co-designed to reduce storage overhead while preserving compatibility with the GPU's execution model. This enables efficient SpMV for unstructured sparsity without specialized hardware units (e.g., tensor cores) or format-specific precomputation. Empirical results show that at sparsity 50%, MACKO is the first approach with significant 1.5x memory reduction and 1.2-1.5x speedup over dense representation. Speedups over other SpMV baselines: 2.8-13.0x over cuSPARSE, 1.9-2.6x over Sputnik, and 2.2-2.5x over DASP. Applied to Llama2-7B pruned with Wanda to sparsity 50%, it delivers 1.5x memory reduction and 1.5x faster inference at fp16 precision. Thanks to MACKO, unstructured pruning at 50% sparsity is now justified in real-world LLM workloads.
Addition is almost all you need: Compressing neural networks with double binary factorization
May 16, 2025Abstract:Binary quantization approaches, which replace weight matrices with binary matrices and substitute costly multiplications with cheaper additions, offer a computationally efficient approach to address the increasing computational and storage requirements of Large Language Models (LLMs). However, the severe quantization constraint ($\pm1$) can lead to significant accuracy degradation. In this paper, we propose Double Binary Factorization (DBF), a novel method that factorizes dense weight matrices into products of two binary (sign) matrices, each accompanied by scaling vectors. DBF preserves the efficiency advantages of binary representations while achieving compression rates that are competitive with or superior to state-of-the-art methods. Specifically, in a 1-bit per weight range, DBF is better than existing binarization approaches. In a 2-bit per weight range, DBF is competitive with the best quantization methods like QuIP\# and QTIP. Unlike most existing compression techniques, which offer limited compression level choices, DBF allows fine-grained control over compression ratios by adjusting the factorization's intermediate dimension. Based on this advantage, we further introduce an algorithm for estimating non-uniform layer-wise compression ratios for DBF, based on previously developed channel pruning criteria. Code available at: https://github.com/usamec/double_binary
Two Sparse Matrices are Better than One: Sparsifying Neural Networks with Double Sparse Factorization
Sep 27, 2024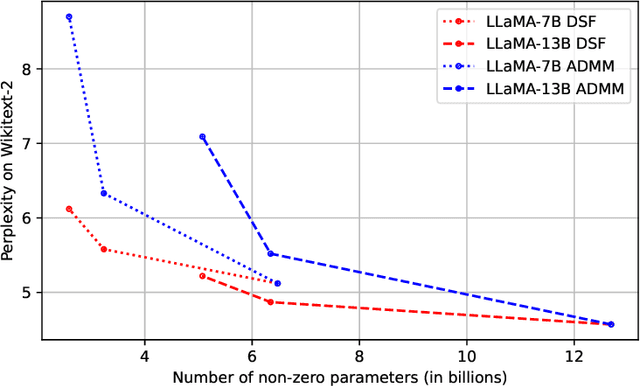
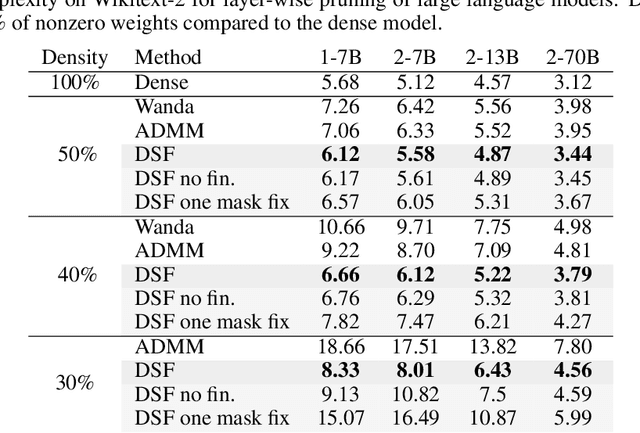
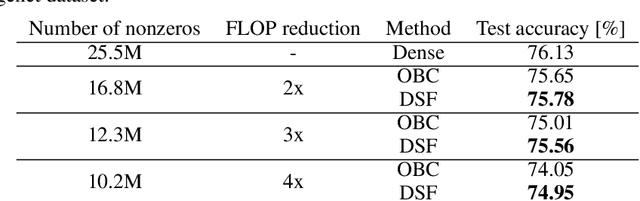
Abstract:Neural networks are often challenging to work with due to their large size and complexity. To address this, various methods aim to reduce model size by sparsifying or decomposing weight matrices, such as magnitude pruning and low-rank or block-diagonal factorization. In this work, we present Double Sparse Factorization (DSF), where we factorize each weight matrix into two sparse matrices. Although solving this problem exactly is computationally infeasible, we propose an efficient heuristic based on alternating minimization via ADMM that achieves state-of-the-art results, enabling unprecedented sparsification of neural networks. For instance, in a one-shot pruning setting, our method can reduce the size of the LLaMA2-13B model by 50% while maintaining better performance than the dense LLaMA2-7B model. We also compare favorably with Optimal Brain Compression, the state-of-the-art layer-wise pruning approach for convolutional neural networks. Furthermore, accuracy improvements of our method persist even after further model fine-tuning. Code available at: https://github.com/usamec/double_sparse.
Fast and Optimal Weight Update for Pruned Large Language Models
Jan 01, 2024Abstract:Pruning large language models (LLMs) is a challenging task due to their enormous size. The primary difficulty is fine-tuning the model after pruning, which is needed to recover the lost performance caused by dropping weights. Recent approaches have either ignored fine-tuning entirely, focusing on efficient pruning criteria, or attempted layer-wise weight updates, preserving the behavior of each layer. However, even layer-wise weight updates can be costly for LLMs, and previous works have resorted to various approximations. In our paper, we propose a fast and optimal weight update algorithm for pruned layers based on the Alternating Direction Method of Multipliers (ADMM). Coupled with a simple iterative pruning mask selection, our algorithm achieves state-of-the-art pruning performance across a wide range of LLMs. Code is available at https://github.com/fmfi-compbio/admm-pruning.
Merging of neural networks
Apr 21, 2022



Abstract:We propose a simple scheme for merging two neural networks trained with different starting initialization into a single one with the same size as the original ones. We do this by carefully selecting channels from each input network. Our procedure might be used as a finalization step after one tries multiple starting seeds to avoid an unlucky one. We also show that training two networks and merging them leads to better performance than training a single network for an extended period of time. Availability: https://github.com/fmfi-compbio/neural-network-merging
Dynamic Pooling Improves Nanopore Base Calling Accuracy
May 16, 2021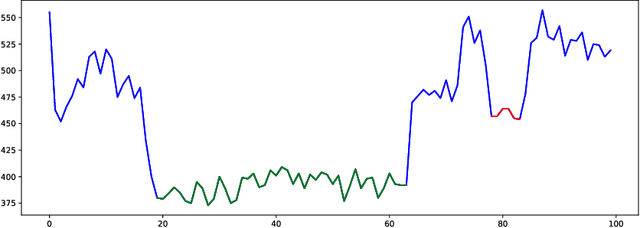
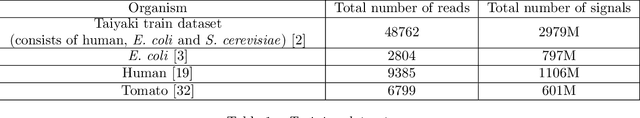
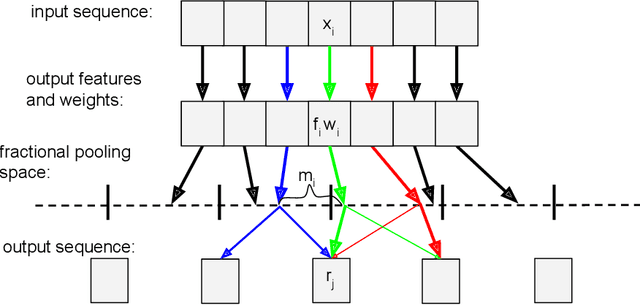
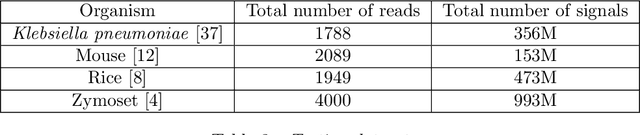
Abstract:In nanopore sequencing, electrical signal is measured as DNA molecules pass through the sequencing pores. Translating these signals into DNA bases (base calling) is a highly non-trivial task, and its quality has a large impact on the sequencing accuracy. The most successful nanopore base callers to date use convolutional neural networks (CNN) to accomplish the task. Convolutional layers in CNNs are typically composed of filters with constant window size, performing best in analysis of signals with uniform speed. However, the speed of nanopore sequencing varies greatly both within reads and between sequencing runs. Here, we present dynamic pooling, a novel neural network component, which addresses this problem by adaptively adjusting the pooling ratio. To demonstrate the usefulness of dynamic pooling, we developed two base callers: Heron and Osprey. Heron improves the accuracy beyond the experimental high-accuracy base caller Bonito developed by Oxford Nanopore. Osprey is a fast base caller that can compete in accuracy with Guppy high-accuracy mode, but does not require GPU acceleration and achieves a near real-time speed on common desktop CPUs. Availability: https://github.com/fmfi-compbio/osprey, https://github.com/fmfi-compbio/heron Keywords: nanopore sequencing, base calling, convolutional neural networks, pooling
Nanopore Base Calling on the Edge
Nov 09, 2020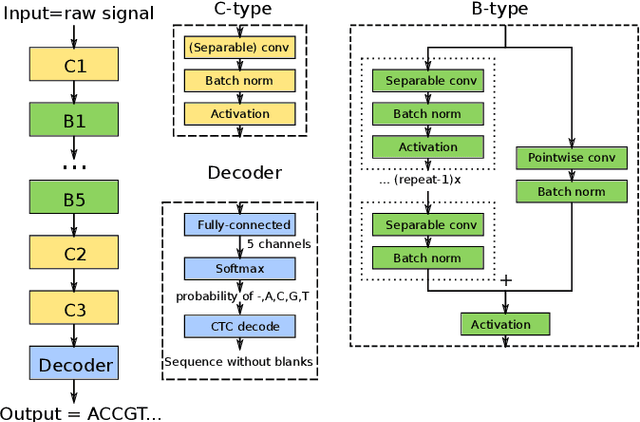
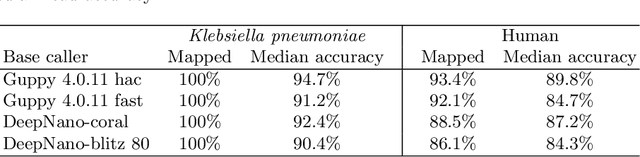
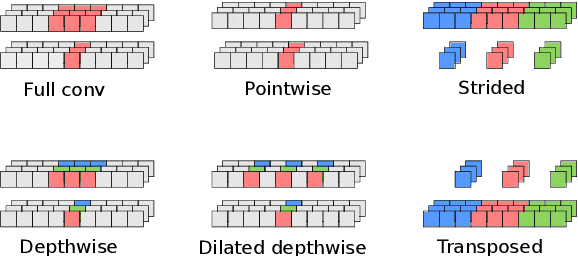
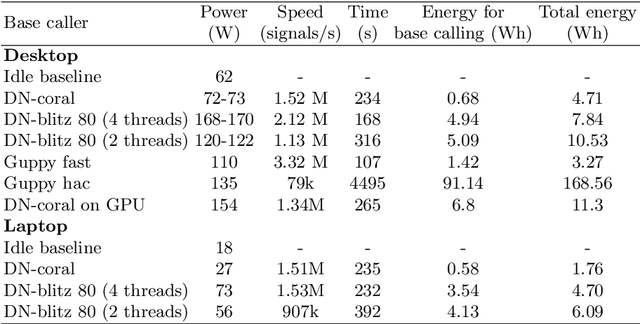
Abstract:We developed a new base caller DeepNano-coral for nanopore sequencing, which is optimized to run on the Coral Edge Tensor Processing Unit, a small USB-attached hardware accelerator. To achieve this goal, we have designed new versions of two key components used in convolutional neural networks for speech recognition and base calling. In our components, we propose a new way of factorization of a full convolution into smaller operations, which decreases memory access operations, memory access being a bottleneck on this device. DeepNano-coral achieves real-time base calling during sequencing with the accuracy slightly better than the fast mode of the Guppy base caller and is extremely energy efficient, using only 10W of power. Availability: https://github.com/fmfi-compbio/coral-basecaller
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge