Violeta Chang
AST-n: A Fast Sampling Approach for Low-Dose CT Reconstruction using Diffusion Models
Aug 13, 2025Abstract:Low-dose CT (LDCT) protocols reduce radiation exposure but increase image noise, compromising diagnostic confidence. Diffusion-based generative models have shown promise for LDCT denoising by learning image priors and performing iterative refinement. In this work, we introduce AST-n, an accelerated inference framework that initiates reverse diffusion from intermediate noise levels, and integrate high-order ODE solvers within conditioned models to further reduce sampling steps. We evaluate two acceleration paradigms--AST-n sampling and standard scheduling with high-order solvers -- on the Low Dose CT Grand Challenge dataset, covering head, abdominal, and chest scans at 10-25 % of standard dose. Conditioned models using only 25 steps (AST-25) achieve peak signal-to-noise ratio (PSNR) above 38 dB and structural similarity index (SSIM) above 0.95, closely matching standard baselines while cutting inference time from ~16 seg to under 1 seg per slice. Unconditional sampling suffers substantial quality loss, underscoring the necessity of conditioning. We also assess DDIM inversion, which yields marginal PSNR gains at the cost of doubling inference time, limiting its clinical practicality. Our results demonstrate that AST-n with high-order samplers enables rapid LDCT reconstruction without significant loss of image fidelity, advancing the feasibility of diffusion-based methods in clinical workflows.
RaViTT: Random Vision Transformer Tokens
Jun 19, 2023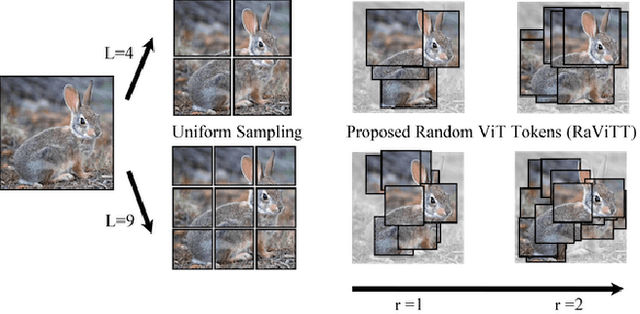
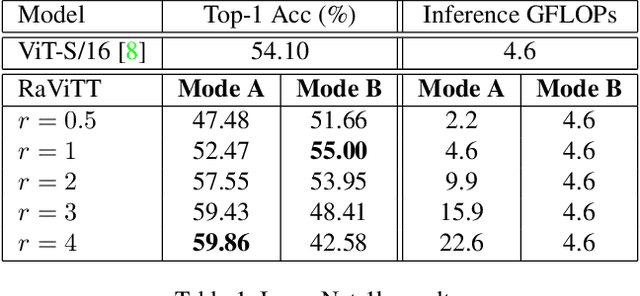
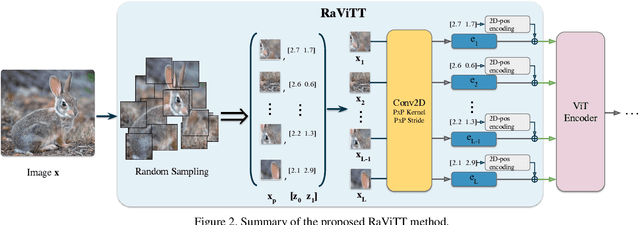
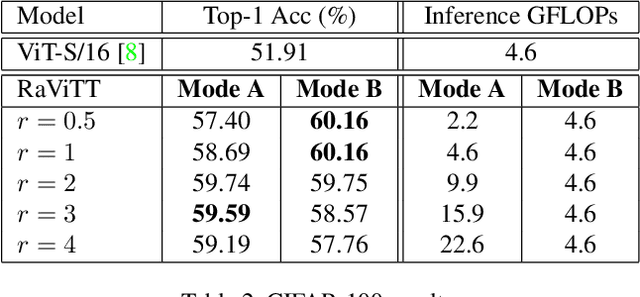
Abstract:Vision Transformers (ViTs) have successfully been applied to image classification problems where large annotated datasets are available. On the other hand, when fewer annotations are available, such as in biomedical applications, image augmentation techniques like introducing image variations or combinations have been proposed. However, regarding ViT patch sampling, less has been explored outside grid-based strategies. In this work, we propose Random Vision Transformer Tokens (RaViTT), a random patch sampling strategy that can be incorporated into existing ViTs. We experimentally evaluated RaViTT for image classification, comparing it with a baseline ViT and state-of-the-art (SOTA) augmentation techniques in 4 datasets, including ImageNet-1k and CIFAR-100. Results show that RaViTT increases the accuracy of the baseline in all datasets and outperforms the SOTA augmentation techniques in 3 out of 4 datasets by a significant margin +1.23% to +4.32%. Interestingly, RaViTT accuracy improvements can be achieved even with fewer tokens, thus reducing the computational load of any ViT model for a given accuracy value.
Gold-standard of HER2 breast cancer biopsies using supervised learning based on multiple pathologist annotations
Nov 09, 2022Abstract:Breast cancer is one of the most common cancer in women around the world. For diagnosis, pathologists evaluate biomarkers such as HER2 protein using immunohistochemistry over tissue extracted by a biopsy. Through microscopic inspection, this assessment estimates the intensity and integrity of the membrane cells' staining and scores the sample as 0, 1+, 2+, or 3+: a subjective decision that depends on the interpretation of the pathologist. This paper presents the preliminary data analysis of the annotations of three pathologists over the same set of samples obtained using 20x magnification and including $1,252$ non-overlapping biopsy patches. We evaluate the intra- and inter-expert variability achieving substantial and moderate agreement, respectively, according to Fleiss' Kappa coefficient, as a previous stage towards a generation of a HER2 breast cancer biopsy gold-standard using supervised learning from multiple pathologist annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge