Vikram Ravindra
Deconvolving Complex Neuronal Networks into Interpretable Task-Specific Connectomes
Jun 28, 2024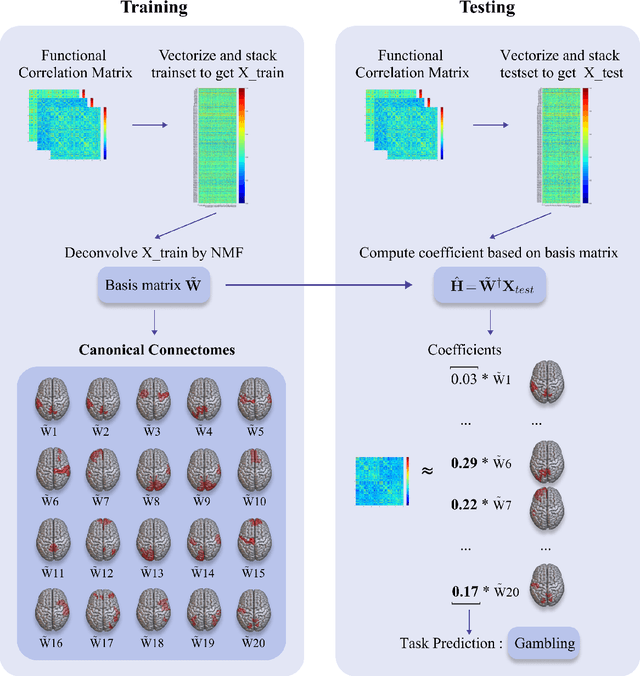
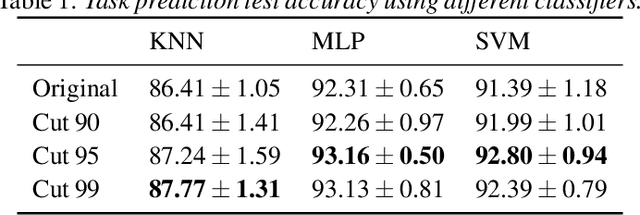
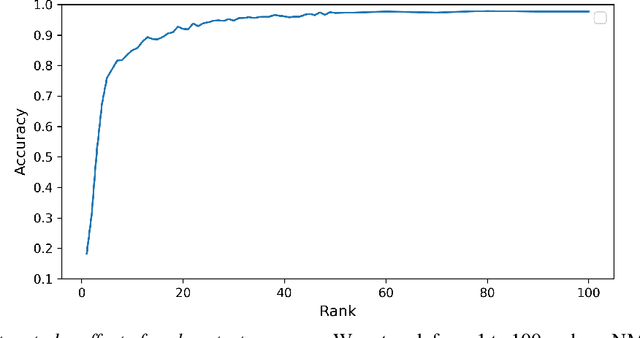
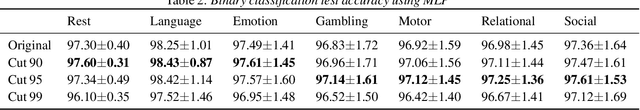
Abstract:Task-specific functional MRI (fMRI) images provide excellent modalities for studying the neuronal basis of cognitive processes. We use fMRI data to formulate and solve the problem of deconvolving task-specific aggregate neuronal networks into a set of basic building blocks called canonical networks, to use these networks for functional characterization, and to characterize the physiological basis of these responses by mapping them to regions of the brain. Our results show excellent task-specificity of canonical networks, i.e., the expression of a small number of canonical networks can be used to accurately predict tasks; generalizability across cohorts, i.e., canonical networks are conserved across diverse populations, studies, and acquisition protocols; and that canonical networks have strong anatomical and physiological basis. From a methods perspective, the problem of identifying these canonical networks poses challenges rooted in the high dimensionality, small sample size, acquisition variability, and noise. Our deconvolution technique is based on non-negative matrix factorization (NMF) that identifies canonical networks as factors of a suitably constructed matrix. We demonstrate that our method scales to large datasets, yields stable and accurate factors, and is robust to noise.
Constructing Compact Brain Connectomes for Individual Fingerprinting
May 22, 2018



Abstract:Recent neuroimaging studies have shown that functional connectomes are unique to individuals, i.e., two distinct fMRIs taken over different sessions of the same subject are more similar in terms of their connectomes than those from two different subjects. In this study, we present significant new results that identify specific parts of the connectome that code the unique signatures. We show that a very small part of the connectome (under 100 features from among over 64K total features) is responsible for the signatures. A network of these features is shown to achieve excellent training and test accuracy in matching imaging datasets. We show that these features are statistically significant, robust to perturbations, invariant across populations, and are localized to a small number of structural regions of the brain (12 regions from among 180). We develop an innovative matrix sampling technique to derive computationally efficient and accurate methods for identifying the discriminating sub-connectome and support all of our claims using state of the art statistical tests and computational techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge