Tung Tran
Attention-Gated Graph Convolutions for Extracting Drug Interaction Information from Drug Labels
Nov 04, 2019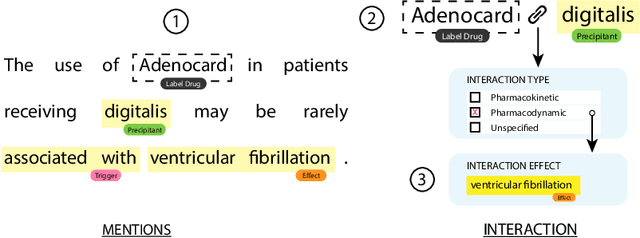
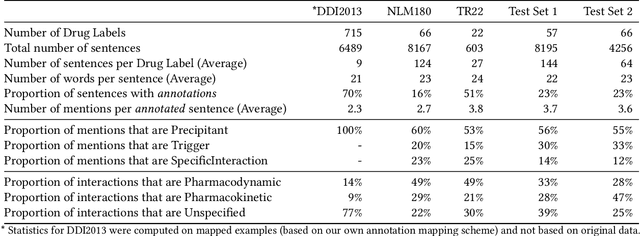
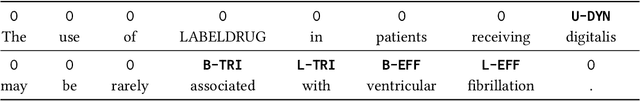
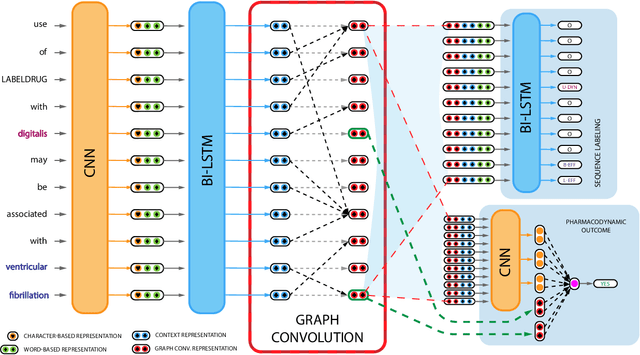
Abstract:Preventable adverse events as a result of medical errors present a growing concern in the healthcare system. As drug-drug interactions (DDIs) may lead to preventable adverse events, being able to extract DDIs from drug labels into a machine-processable form is an important step toward effective dissemination of drug safety information. In this study, we tackle the problem of jointly extracting drugs and their interactions, including interaction outcome, from drug labels. Our deep learning approach entails composing various intermediate representations including sequence and graph based context, where the latter is derived using graph convolutions (GC) with a novel attention-based gating mechanism (holistically called GCA). These representations are then composed in meaningful ways to handle all subtasks jointly. To overcome scarcity in training data, we additionally propose transfer learning by pre-training on related DDI data. Our model is trained and evaluated on the 2018 TAC DDI corpus. Our GCA model in conjunction with transfer learning performs at 39.20% F1 and 26.09% F1 on entity recognition (ER) and relation extraction (RE) respectively on the first official test set and at 45.30% F1 and 27.87% F1 on ER and RE respectively on the second official test set corresponding to an improvement over our prior best results by up to 6 absolute F1 points. After controlling for available training data, our model exhibits state-of-the-art performance by improving over the next comparable best outcome by roughly three F1 points in ER and 1.5 F1 points in RE evaluation across two official test sets.
Neural Metric Learning for Fast End-to-End Relation Extraction
Jun 03, 2019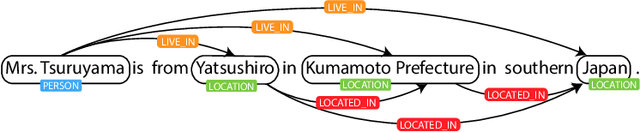
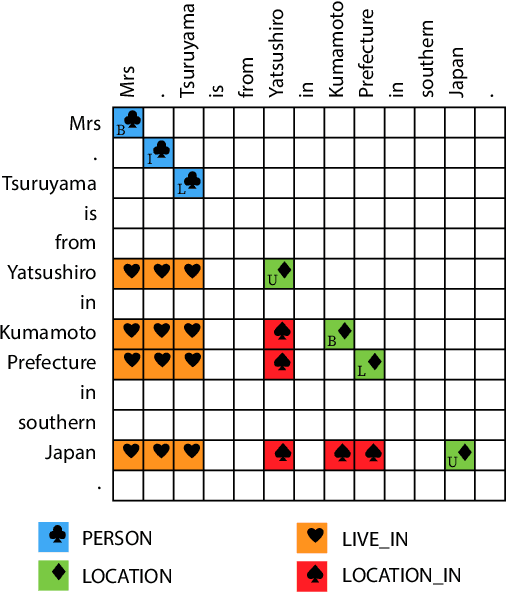
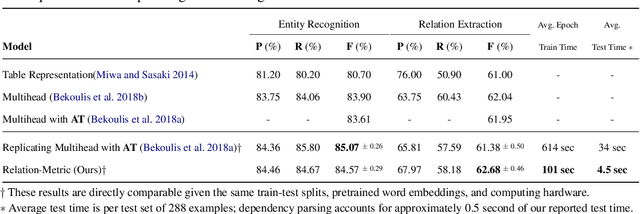
Abstract:Relation extraction (RE) is an indispensable information extraction task in several disciplines. RE models typically assume that named entity recognition (NER) is already performed in a previous step by another independent model. Several recent efforts, under the theme of end-to-end RE, seek to exploit inter-task correlations by modeling both NER and RE tasks jointly. Earlier work in this area commonly reduces the task to a table-filling problem wherein an additional expensive decoding step involving beam search is applied to obtain globally consistent cell labels. In efforts that do not employ table-filling, global optimization in the form of CRFs with Viterbi decoding for the NER component is still necessary for competitive performance. We introduce a novel neural architecture utilizing the table structure, based on repeated applications of 2D convolutions for pooling local dependency and metric-based features, that improves on the state-of-the-art without the need for global optimization. We validate our model on the ADE and CoNLL04 datasets for end-to-end RE and demonstrate $\approx 1\%$ gain (in F-score) over prior best results with training and testing times that are seven to ten times faster --- the latter highly advantageous for time-sensitive end user applications.
A Multi-Task Learning Framework for Extracting Drugs and Their Interactions from Drug Labels
May 17, 2019
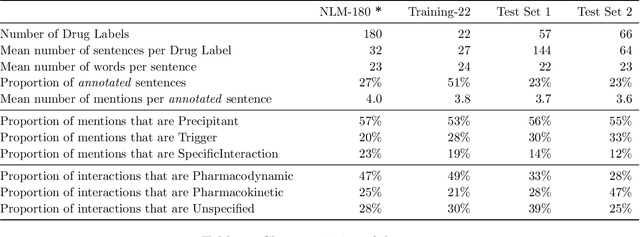
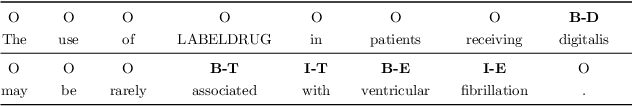
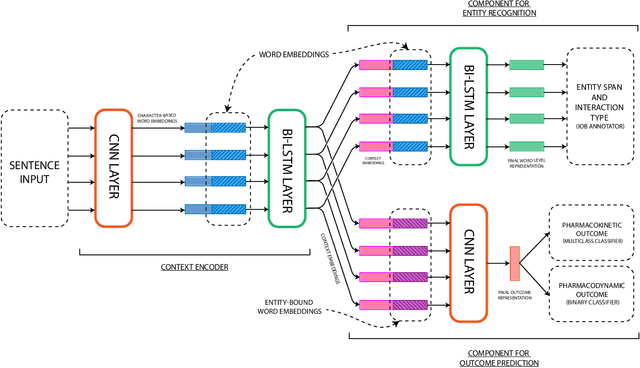
Abstract:Preventable adverse drug reactions as a result of medical errors present a growing concern in modern medicine. As drug-drug interactions (DDIs) may cause adverse reactions, being able to extracting DDIs from drug labels into machine-readable form is an important effort in effectively deploying drug safety information. The DDI track of TAC 2018 introduces two large hand-annotated test sets for the task of extracting DDIs from structured product labels with linkage to standard terminologies. Herein, we describe our approach to tackling tasks one and two of the DDI track, which corresponds to named entity recognition (NER) and sentence-level relation extraction respectively. Namely, our approach resembles a multi-task learning framework designed to jointly model various sub-tasks including NER and interaction type and outcome prediction. On NER, our system ranked second (among eight teams) at 33.00% and 38.25% F1 on Test Sets 1 and 2 respectively. On relation extraction, our system ranked second (among four teams) at 21.59% and 23.55% on Test Sets 1 and 2 respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge