Tugberk Erol
PlutoNet: An Efficient Polyp Segmentation Network
Apr 12, 2022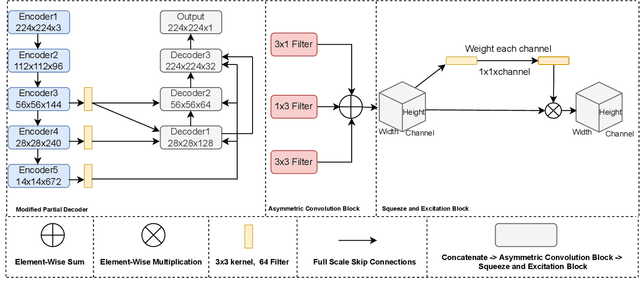
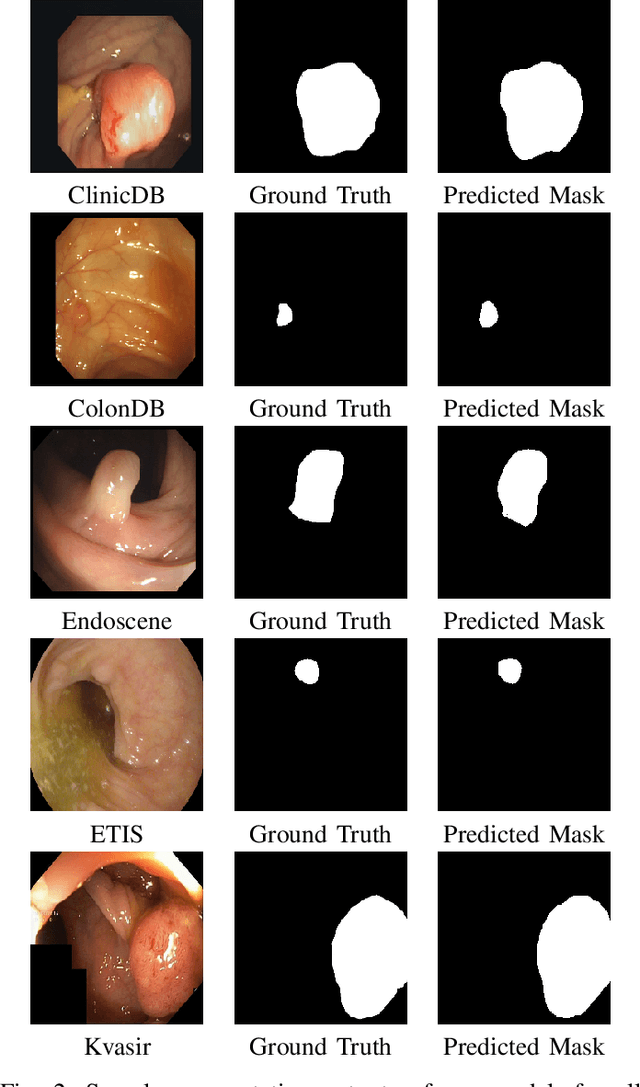
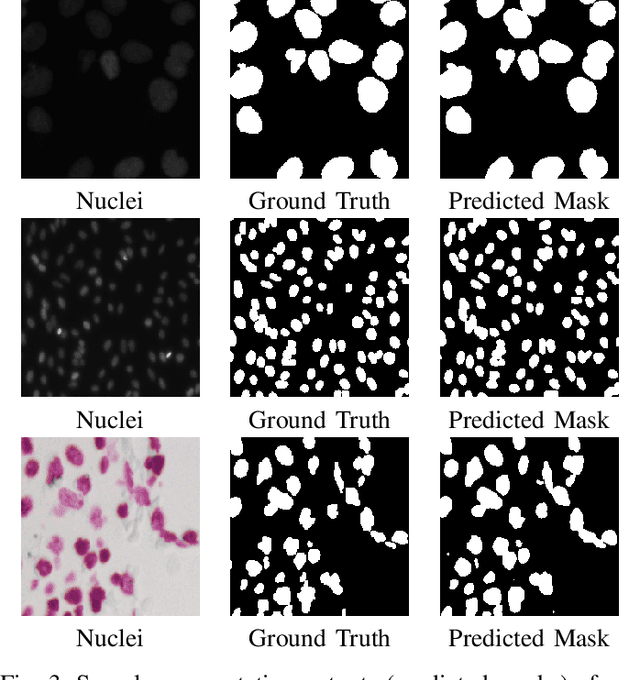
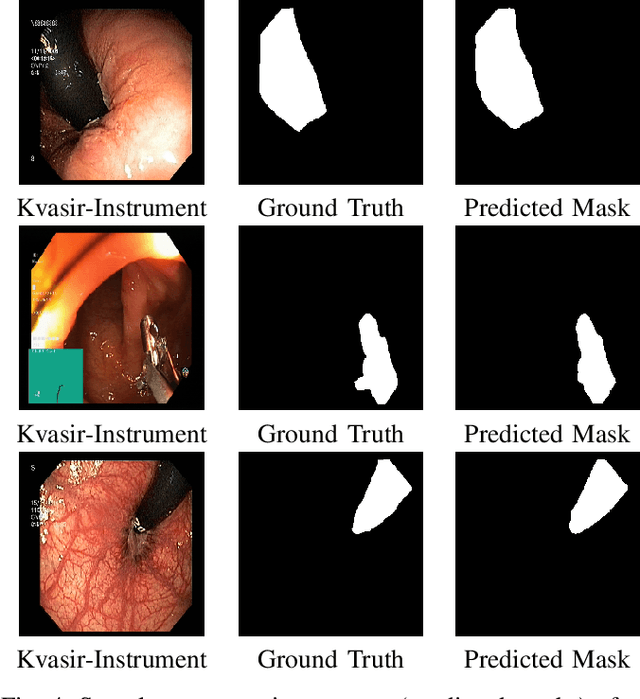
Abstract:Polyps in the colon can turn into cancerous cells if not removed with early intervention. Deep learning models are used to minimize the number of polyps that goes unnoticed by the experts, and to accurately segment the detected polyps during these interventions. Although these models perform well on these tasks, they require too many parameters, which can pose a problem with real-time applications. To address this problem, we propose a novel segmentation model called PlutoNet which requires only 2,626,337 parameters while outperforming state-of-the-art models on multiple medical image segmentation tasks. We use EfficientNetB0 architecture as a backbone and propose the novel \emph{modified partial decoder}, which is a combination of partial decoder and full scale connections, which further reduces the number of parameters required, as well as captures semantic details. We use asymmetric convolutions to handle varying polyp sizes. Finally, we weight each feature map to improve segmentation by using a squeeze and excitation block. In addition to polyp segmentation in colonoscopy, we tested our model on segmentation of nuclei and surgical instruments to demonstrate its generalizability to different medical image segmentation tasks. Our model outperformed the state-of-the-art models with a Dice score of \%92.3 in CVC-ClinicDB dataset and \%89.3 in EndoScene dataset, a Dice score of \%91.93 on the 2018 Data Science Bowl Challenge dataset, and a Dice score of \%94.8 on Kvasir-Instrument dataset. Our experiments and ablation studies show that our model is superior in terms of accuracy, and it is able generalize well to multiple medical segmentation tasks.
An Efficient Polyp Segmentation Network
Mar 08, 2022Abstract:Cancer is a disease that occurs as a result of uncontrolled division and proliferation of cells. The number of cancer cases has been on the rise over the recent years.. Colon cancer is one of the most common types of cancer in the world. Polyps that can be seen in the large intestine can cause cancer if not removed with early intervention. Deep learning and image segmentation techniques are used to minimize the number of polyps that goes unnoticed by the experts during the diagnosis. Although these techniques give good results, they require too many parameters. We propose a new model to solve this problem. Our proposed model includes less parameters as well as outperforming the success of the state of the art models. In the proposed model, a partial decoder is used to reduce the number of parameters while maintaning success. EfficientNetB0, which gives successfull results as well as requiring few parameters, is used in the encoder part. Since polyps have variable aspect and aspect ratios, an asymetric convolution block was used instead of using classic convolution block. Kvasir and CVC-ClinicDB datasets were seperated as training, validation and testing, and CVC-ColonDB, ETIS and Endoscene datasets were used for testing. According to the dice metric, our model had the best results with %71.8 in the ColonDB test dataset, %89.3 in the EndoScene test dataset and %74.8 in the ETIS test dataset. Our model requires a total of 2.626.337 parameters. When we compare it in the literature, according to similar studies, the model that requires the least parameters is U-Net++ with 9.042.177 parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge