Trung Hoang Le
An LLM + ASP Workflow for Joint Entity-Relation Extraction
Aug 18, 2025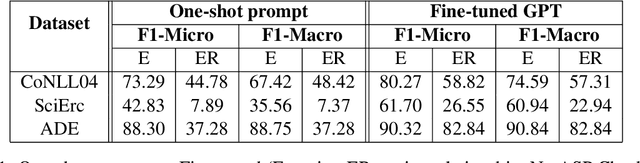
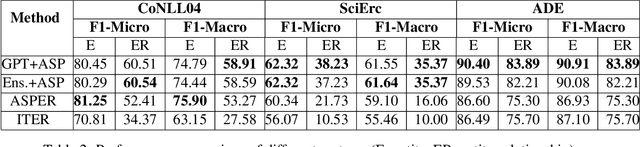
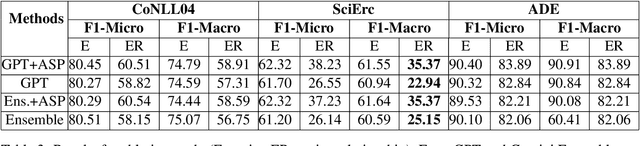
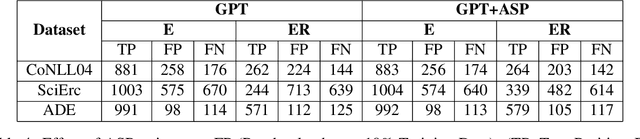
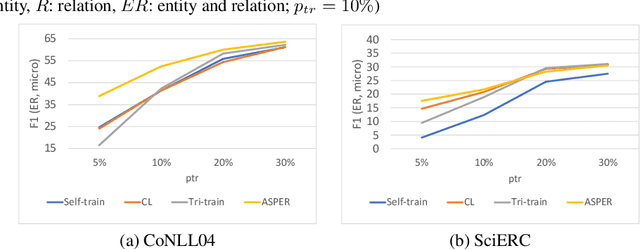
Abstract:Joint entity-relation extraction (JERE) identifies both entities and their relationships simultaneously. Traditional machine-learning based approaches to performing this task require a large corpus of annotated data and lack the ability to easily incorporate domain specific information in the construction of the model. Therefore, creating a model for JERE is often labor intensive, time consuming, and elaboration intolerant. In this paper, we propose harnessing the capabilities of generative pretrained large language models (LLMs) and the knowledge representation and reasoning capabilities of Answer Set Programming (ASP) to perform JERE. We present a generic workflow for JERE using LLMs and ASP. The workflow is generic in the sense that it can be applied for JERE in any domain. It takes advantage of LLM's capability in natural language understanding in that it works directly with unannotated text. It exploits the elaboration tolerant feature of ASP in that no modification of its core program is required when additional domain specific knowledge, in the form of type specifications, is found and needs to be used. We demonstrate the usefulness of the proposed workflow through experiments with limited training data on three well-known benchmarks for JERE. The results of our experiments show that the LLM + ASP workflow is better than state-of-the-art JERE systems in several categories with only 10\% of training data. It is able to achieve a 2.5 times (35\% over 15\%) improvement in the Relation Extraction task for the SciERC corpus, one of the most difficult benchmarks.
ASPER: Answer Set Programming Enhanced Neural Network Models for Joint Entity-Relation Extraction
May 24, 2023
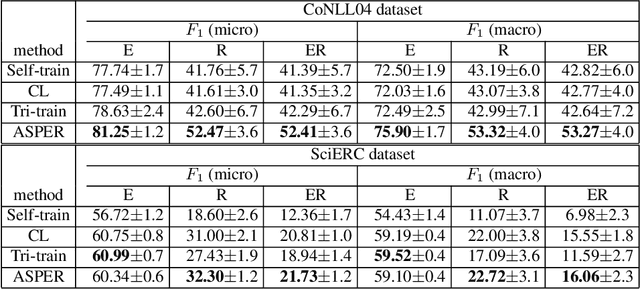
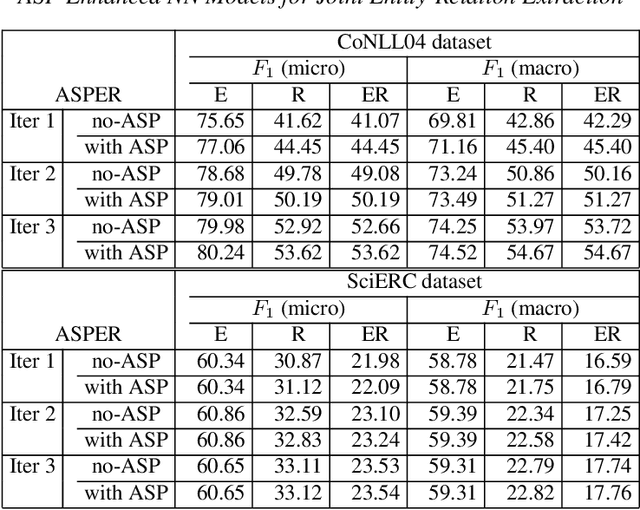
Abstract:A plethora of approaches have been proposed for joint entity-relation (ER) extraction. Most of these methods largely depend on a large amount of manually annotated training data. However, manual data annotation is time consuming, labor intensive, and error prone. Human beings learn using both data (through induction) and knowledge (through deduction). Answer Set Programming (ASP) has been a widely utilized approach for knowledge representation and reasoning that is elaboration tolerant and adept at reasoning with incomplete information. This paper proposes a new approach, ASP-enhanced Entity-Relation extraction (ASPER), to jointly recognize entities and relations by learning from both data and domain knowledge. In particular, ASPER takes advantage of the factual knowledge (represented as facts in ASP) and derived knowledge (represented as rules in ASP) in the learning process of neural network models. We have conducted experiments on two real datasets and compare our method with three baselines. The results show that our ASPER model consistently outperforms the baselines.
Attention-based Multi-Input Deep Learning Architecture for Biological Activity Prediction: An Application in EGFR Inhibitors
Jun 12, 2019
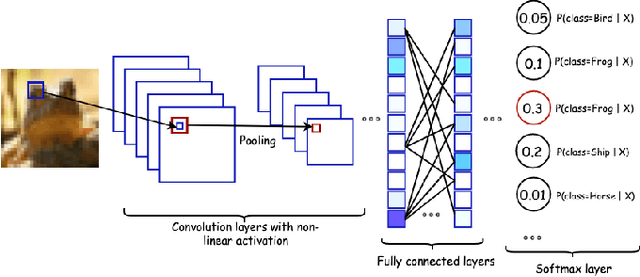
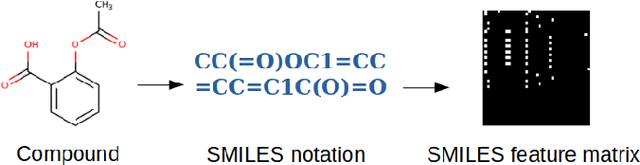
Abstract:Machine learning and deep learning have gained popularity and achieved immense success in Drug discovery in recent decades. Historically, machine learning and deep learning models were trained on either structural data or chemical properties by separated model. In this study, we proposed an architecture training simultaneously both type of data in order to improve the overall performance. Given the molecular structure in the form of SMILES notation and their label, we generated the SMILES-based feature matrix and molecular descriptors. These data was trained on an deep learning model which was also integrated with the Attention mechanism to facilitate training and interpreting. Experiments showed that our model could raise the performance of model. With the maximum MCC 0.56 and AUC 91% by cross-validation on EGFR inhibitors dataset, our architecture was outperforming the referring model. We also successfully integrated Attention mechanism into our model, which helped to interpret the contribution of chemical structures on bioactivity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge