Tomislav Šmuc
Multi-view redescription mining using tree-based multi-target prediction models
Jun 22, 2020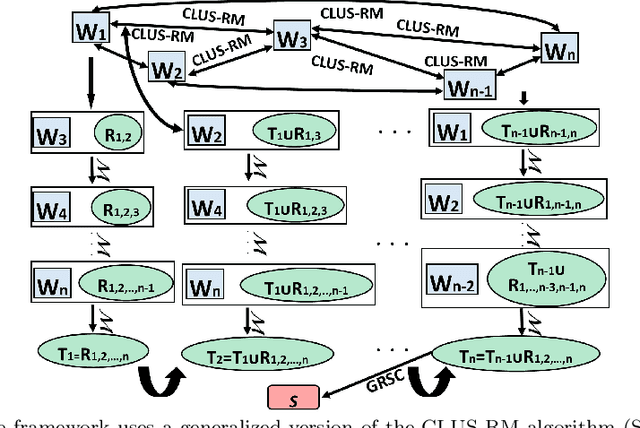
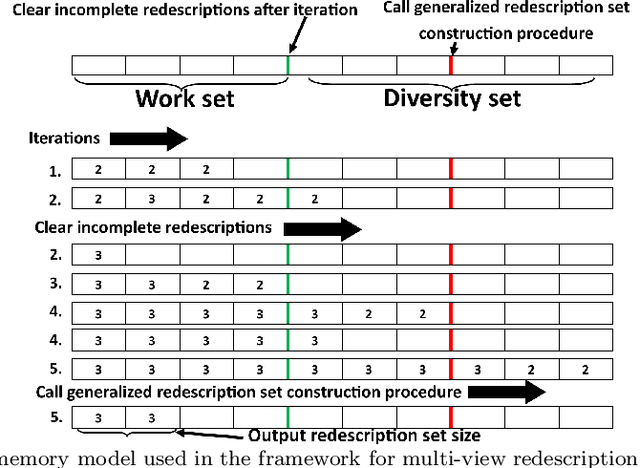
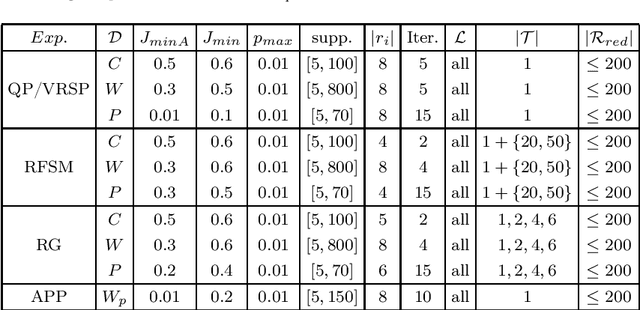
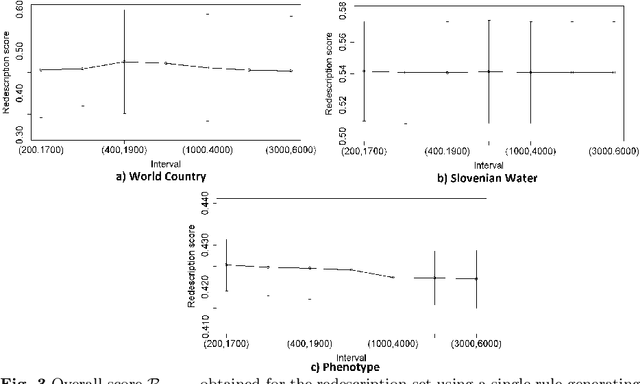
Abstract:The task of redescription mining is concerned with re-describing different subsets of entities contained in a dataset and revealing non-trivial associations between different subsets of attributes, called views. This interesting and challenging task is encountered in different scientific fields, and is addressed by a number of approaches that obtain redescriptions and allow for the exploration and analysis of attribute associations. The main limitation of existing approaches to this task is their inability to use more than two views. Our work alleviates this drawback. We present a memory efficient, extensible multi-view redescription mining framework that can be used to relate multiple, i.e. more than two views, disjoint sets of attributes describing one set of entities. The framework includes: a) the use of random forest of Predictive Clustering trees, with and without random output selection, and random forests of Extra Predictive Clustering trees, b) using Extra Predictive Clustering trees as a main rule generation mechanism in the framework and c) using random view subset projections. We provide multiple performance analyses of the proposed framework and demonstrate its usefulness in increasing the understanding of different machine learning models, which has become a topic of growing importance in machine learning and especially in the field of computer science called explainable data science.
Using Redescription Mining to Relate Clinical and Biological Characteristics of Cognitively Impaired and Alzheimer's Disease Patients
Nov 14, 2017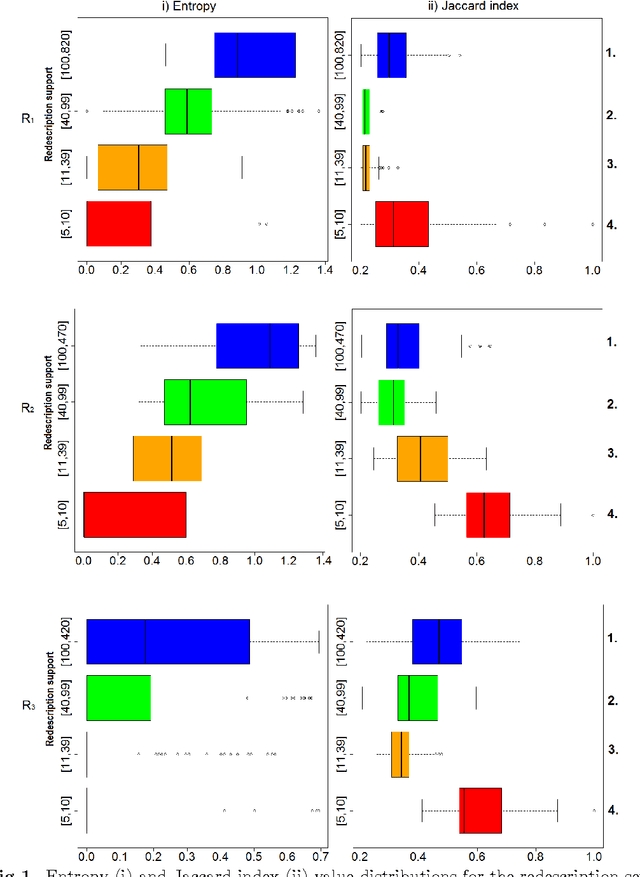
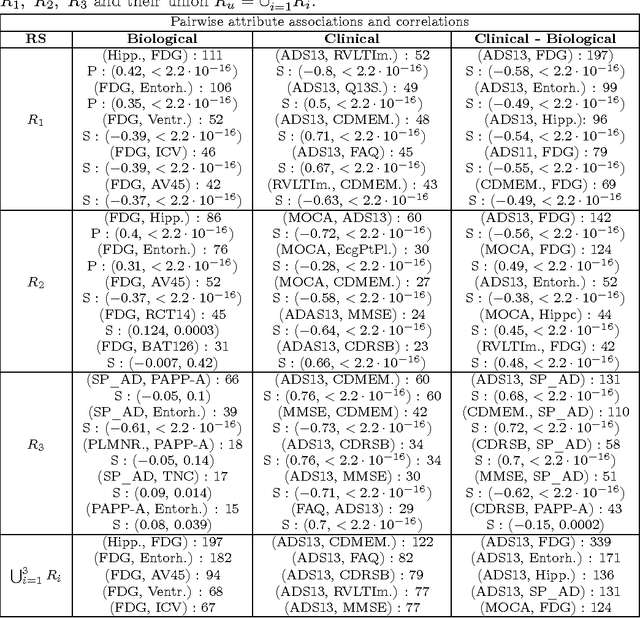
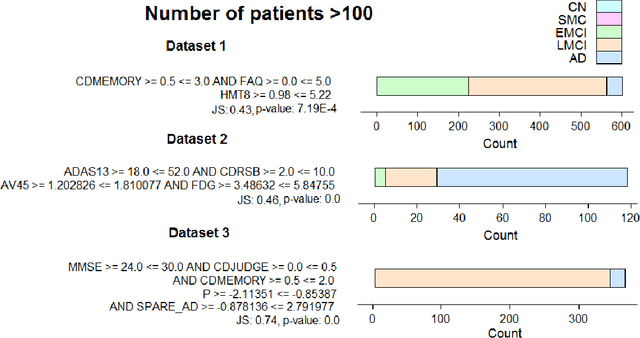
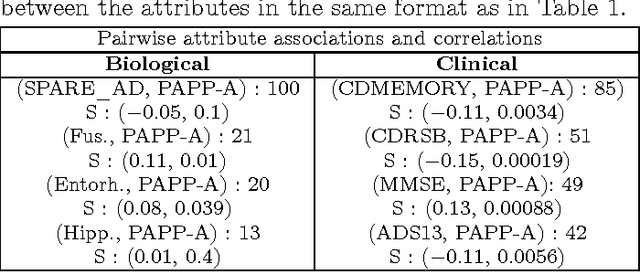
Abstract:We used redescription mining to find interpretable rules revealing associations between those determinants that provide insights about the Alzheimer's disease (AD). We extended the CLUS-RM redescription mining algorithm to a constraint-based redescription mining (CBRM) setting, which enables several modes of targeted exploration of specific, user-constrained associations. Redescription mining enabled finding specific constructs of clinical and biological attributes that describe many groups of subjects of different size, homogeneity and levels of cognitive impairment. We confirmed some previously known findings. However, in some instances, as with the attributes: testosterone, the imaging attribute Spatial Pattern of Abnormalities for Recognition of Early AD, as well as the levels of leptin and angiopoietin-2 in plasma, we corroborated previously debatable findings or provided additional information about these variables and their association with AD pathogenesis. Applying redescription mining on ADNI data resulted with the discovery of one largely unknown attribute: the Pregnancy-Associated Protein-A (PAPP-A), which we found highly associated with cognitive impairment in AD. Statistically significant correlations (p <= 0.01) were found between PAPP-A and various different clinical tests. The high importance of this finding lies in the fact that PAPP-A is a metalloproteinase, known to cleave insulin-like growth factor binding proteins. Since it also shares similar substrates with A Disintegrin and the Metalloproteinase family of enzymes that act as {\alpha}-secretase to physiologically cleave amyloid precursor protein (APP) in the non-amyloidogenic pathway, it could be directly involved in the metabolism of APP very early during the disease course. Therefore, further studies should investigate the role of PAPP-A in the development of AD more thoroughly.
A framework for redescription set construction
Dec 19, 2016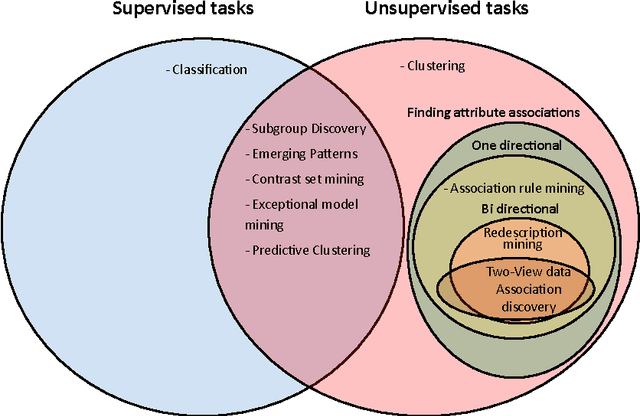
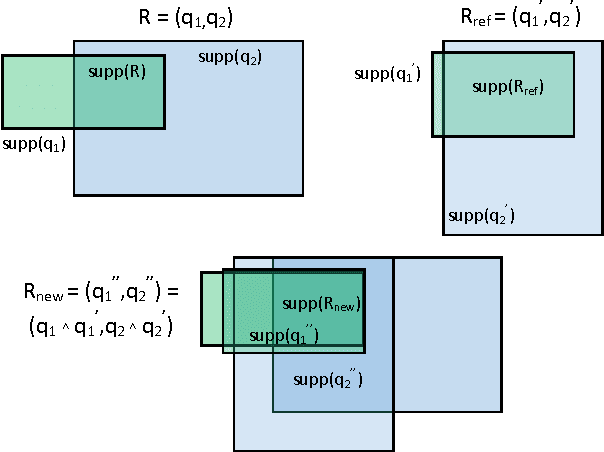
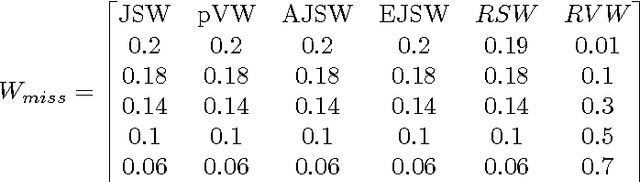
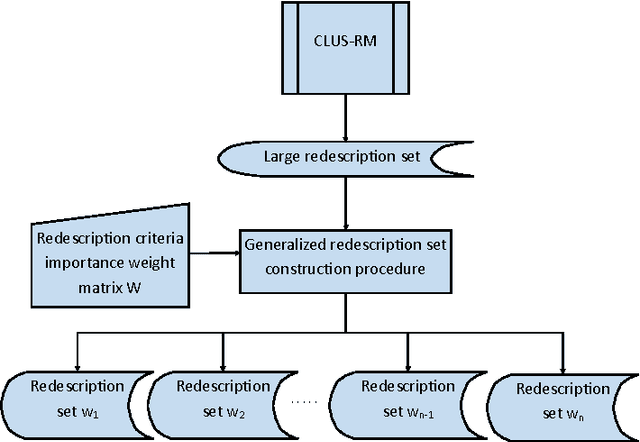
Abstract:Redescription mining is a field of knowledge discovery that aims at finding different descriptions of similar subsets of instances in the data. These descriptions are represented as rules inferred from one or more disjoint sets of attributes, called views. As such, they support knowledge discovery process and help domain experts in formulating new hypotheses or constructing new knowledge bases and decision support systems. In contrast to previous approaches that typically create one smaller set of redescriptions satisfying a pre-defined set of constraints, we introduce a framework that creates large and heterogeneous redescription set from which user/expert can extract compact sets of differing properties, according to its own preferences. Construction of large and heterogeneous redescription set relies on CLUS-RM algorithm and a novel, conjunctive refinement procedure that facilitates generation of larger and more accurate redescription sets. The work also introduces the variability of redescription accuracy when missing values are present in the data, which significantly extends applicability of the method. Crucial part of the framework is the redescription set extraction based on heuristic multi-objective optimization procedure that allows user to define importance levels towards one or more redescription quality criteria. We provide both theoretical and empirical comparison of the novel framework against current state of the art redescription mining algorithms and show that it represents more efficient and versatile approach for mining redescriptions from data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge