Tom Velez
Identification of Pediatric Sepsis Subphenotypes for Enhanced Machine Learning Predictive Performance: A Latent Profile Analysis
Aug 23, 2019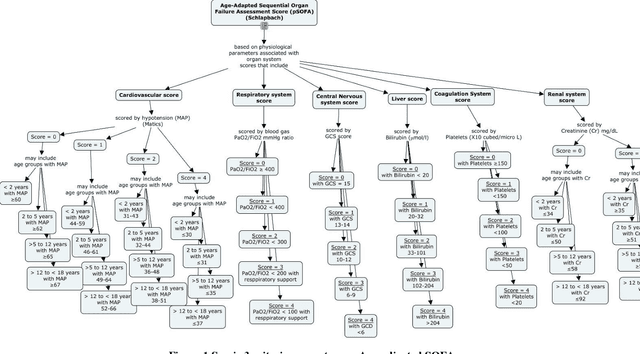
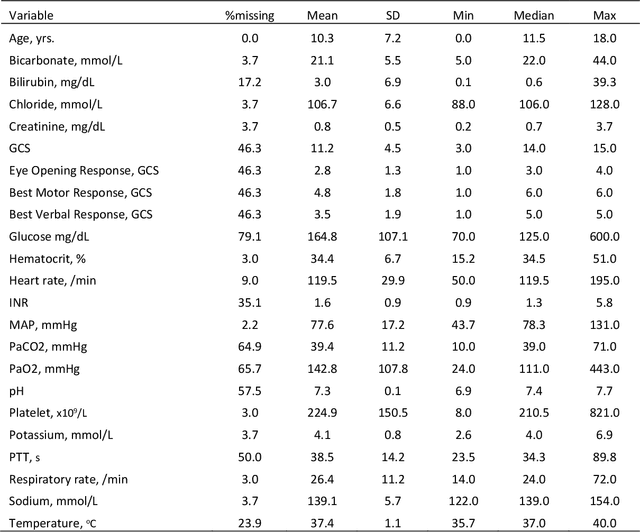
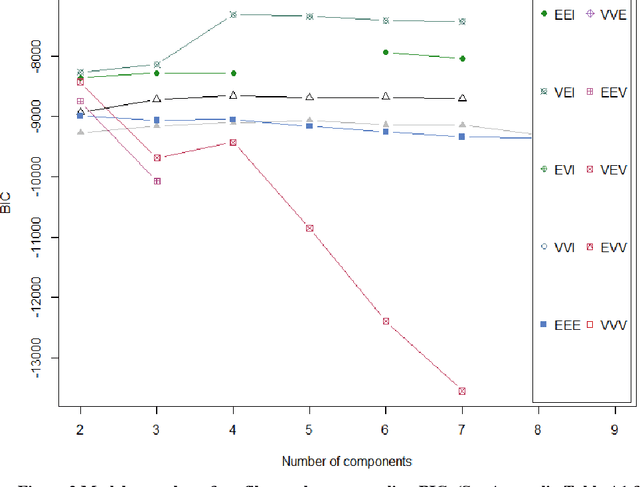
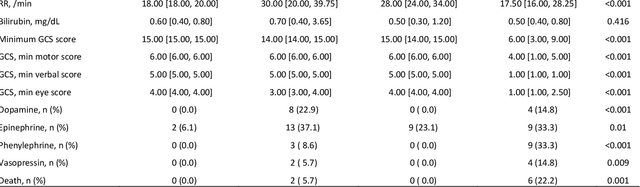
Abstract:Background: While machine learning (ML) models are rapidly emerging as promising screening tools in critical care medicine, the identification of homogeneous subphenotypes within populations with heterogeneous conditions such as pediatric sepsis may facilitate attainment of high-predictive performance of these prognostic algorithms. This study is aimed to identify subphenotypes of pediatric sepsis and demonstrate the potential value of partitioned data/subtyping-based training. Methods: This was a retrospective study of clinical data extracted from medical records of 6,446 pediatric patients that were admitted at a major hospital system in the DC area. Vitals and labs associated with patients meeting the diagnostic criteria for sepsis were used to perform latent profile analysis. Modern ML algorithms were used to explore the predictive performance benefits of reduced training data heterogeneity via label profiling. Results: In total 134 (2.1%) patients met the diagnostic criteria for sepsis in this cohort and latent profile analysis identified four profiles/subphenotypes of pediatric sepsis. Profiles 1 and 3 had the lowest mortality and included pediatric patients from different age groups. Profile 2 were characterized by respiratory dysfunction; profile 4 by neurological dysfunction and highest mortality rate (22.2%). Machine learning experiments comparing the predictive performance of models derived without training data profiling against profile targeted models suggest statistically significant improved performance of prediction can be obtained. For example, area under ROC curve (AUC) obtained to predict profile 4 with 24-hour data (AUC = .998, p < .0001) compared favorably with the AUC obtained from the model considering all profiles as a single homogeneous group (AUC = .918) with 24-hour data.
Using Latent Class Analysis to Identify ARDS Sub-phenotypes for Enhanced Machine Learning Predictive Performance
Mar 28, 2019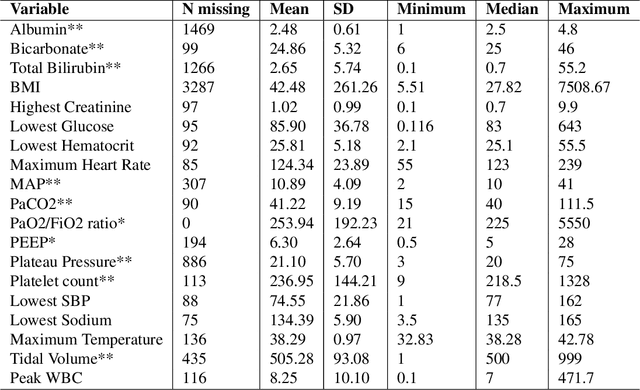

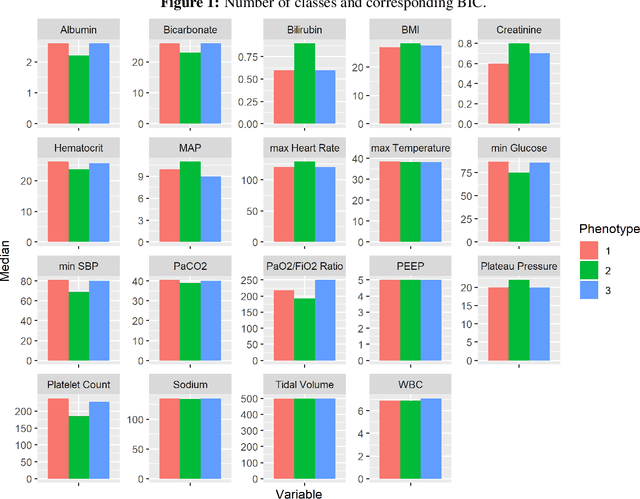
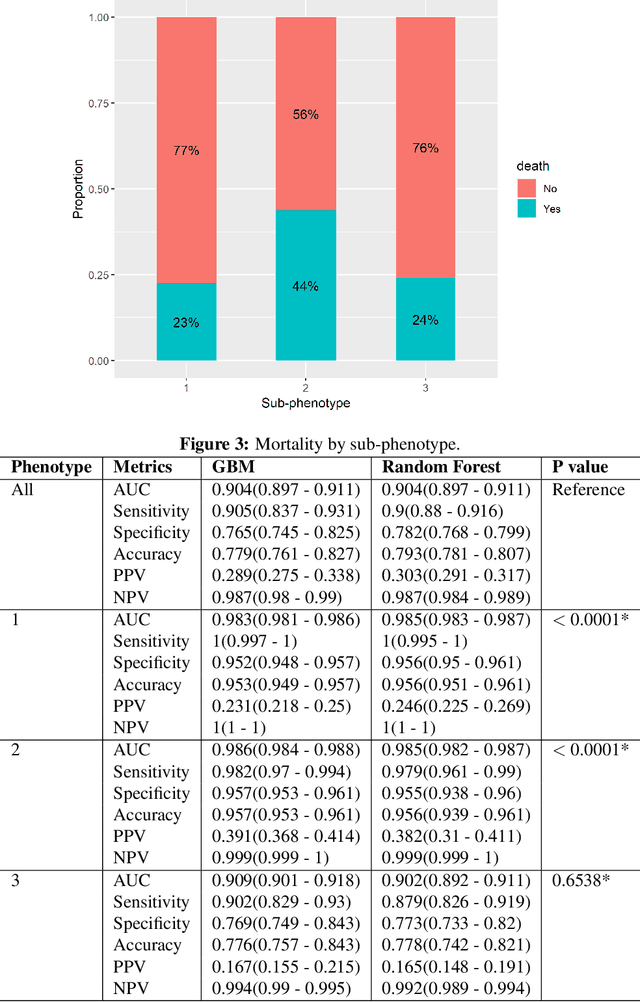
Abstract:In this work, we utilize Machine Learning for early recognition of patients at high risk of acute respiratory distress syndrome (ARDS), which is critical for successful prevention strategies for this devastating syndrome. The difficulty in early ARDS recognition stems from its complex and heterogenous nature. In this study, we integrate knowledge of the heterogeneity of ARDS patients into predictive model building. Using MIMIC-III data, we first apply latent class analysis (LCA) to identify homogeneous sub-groups in the ARDS population, and then build predictive models on the partitioned data. The results indicate that significantly improved performances of prediction can be obtained for two of the three identified sub-phenotypes of ARDS. Experiments suggests that identifying sub-phenotypes is beneficial for building predictive model for ARDS.
Semantically Enhanced Dynamic Bayesian Network for Detecting Sepsis Mortality Risk in ICU Patients with Infection
Jun 26, 2018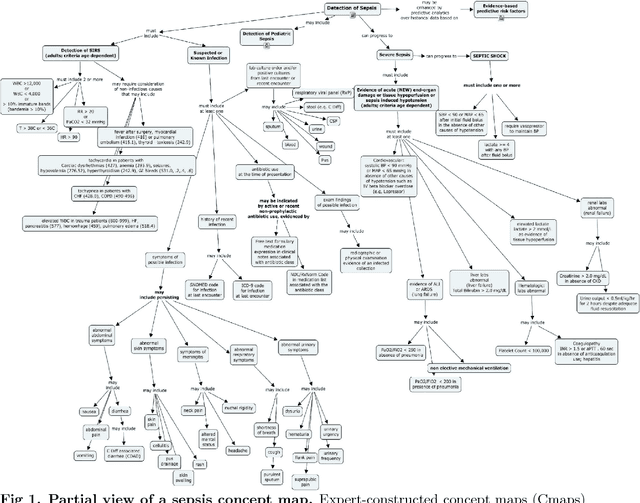
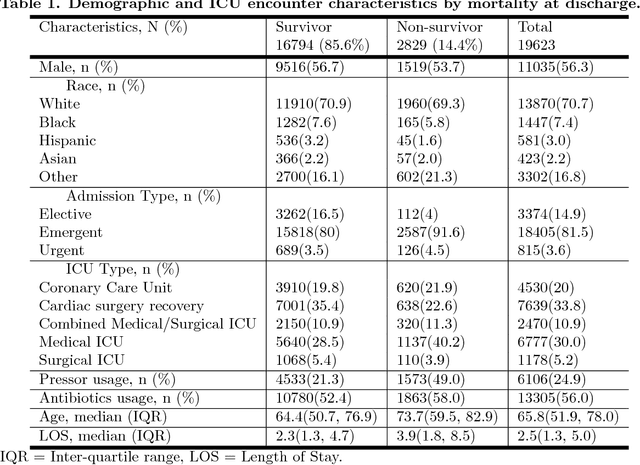
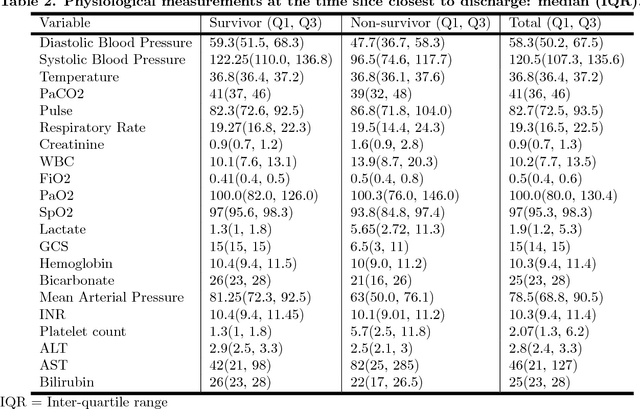
Abstract:Although timely sepsis diagnosis and prompt interventions in Intensive Care Unit (ICU) patients are associated with reduced mortality, early clinical recognition is frequently impeded by non-specific signs of infection and failure to detect signs of sepsis-induced organ dysfunction in a constellation of dynamically changing physiological data. The goal of this work is to identify patient at risk of life-threatening sepsis utilizing a data-centered and machine learning-driven approach. We derive a mortality risk predictive dynamic Bayesian network (DBN) guided by a customized sepsis knowledgebase and compare the predictive accuracy of the derived DBN with the Sepsis-related Organ Failure Assessment (SOFA) score, the Quick SOFA (qSOFA) score, the Simplified Acute Physiological Score (SAPS-II) and the Modified Early Warning Score (MEWS) tools. A customized sepsis ontology was used to derive the DBN node structure and semantically characterize temporal features derived from both structured physiological data and unstructured clinical notes. We assessed the performance in predicting mortality risk of the DBN predictive model and compared performance to other models using Receiver Operating Characteristic (ROC) curves, area under curve (AUROC), calibration curves, and risk distributions. The derived dataset consists of 24,506 ICU stays from 19,623 patients with evidence of suspected infection, with 2,829 patients deceased at discharge. The DBN AUROC was found to be 0.91, which outperformed the SOFA (0.843), qSOFA (0.66), MEWS (0.73), and SAPS-II (0.77) scoring tools. Continuous Net Reclassification Index and Integrated Discrimination Improvement analysis supported the superiority DBN. Compared with conventional rule-based risk scoring tools, the sepsis knowledgebase-driven DBN algorithm offers improved performance for predicting mortality of infected patients in ICUs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge