Tiep Huu Vu
Signal Classification under structure sparsity constraints
Dec 28, 2018
Abstract:Object Classification is a key direction of research in signal and image processing, computer vision and artificial intelligence. The goal is to come up with algorithms that automatically analyze images and put them in predefined categories. This dissertation focuses on the theory and application of sparse signal processing and learning algorithms for image processing and computer vision, especially object classification problems. A key emphasis of this work is to formulate novel optimization problems for learning dictionary and structured sparse representations. Tractable solutions are proposed subsequently for the corresponding optimization problems. An important goal of this dissertation is to demonstrate the wide applications of these algorithmic tools for real-world applications. To that end, we explored important problems in the areas of: 1. Medical imaging: histopathological images acquired from mammalian tissues, human breast tissues, and human brain tissues. 2. Low-frequency (UHF to L-band) ultra-wideband (UWB) synthetic aperture radar: detecting bombs and mines buried under rough surfaces. 3. General object classification: face, flowers, objects, dogs, indoor scenes, etc.
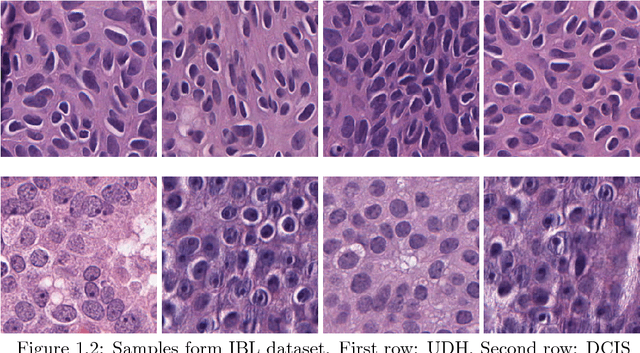
Histopathological Image Classification using Discriminative Feature-oriented Dictionary Learning
Mar 29, 2016



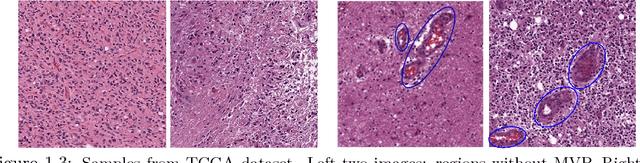
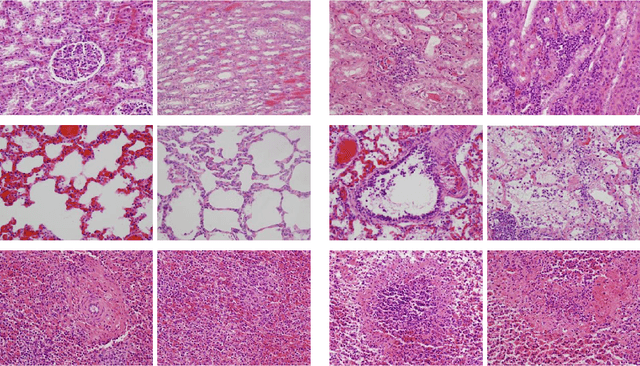
Abstract:In histopathological image analysis, feature extraction for classification is a challenging task due to the diversity of histology features suitable for each problem as well as presence of rich geometrical structures. In this paper, we propose an automatic feature discovery framework via learning class-specific dictionaries and present a low-complexity method for classification and disease grading in histopathology. Essentially, our Discriminative Feature-oriented Dictionary Learning (DFDL) method learns class-specific dictionaries such that under a sparsity constraint, the learned dictionaries allow representing a new image sample parsimoniously via the dictionary corresponding to the class identity of the sample. At the same time, the dictionary is designed to be poorly capable of representing samples from other classes. Experiments on three challenging real-world image databases: 1) histopathological images of intraductal breast lesions, 2) mammalian kidney, lung and spleen images provided by the Animal Diagnostics Lab (ADL) at Pennsylvania State University, and 3) brain tumor images from The Cancer Genome Atlas (TCGA) database, reveal the merits of our proposal over state-of-the-art alternatives. {Moreover, we demonstrate that DFDL exhibits a more graceful decay in classification accuracy against the number of training images which is highly desirable in practice where generous training is often not available
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge