Tiancheng Xia
Automated Blood Cell Detection and Counting via Deep Learning for Microfluidic Point-of-Care Medical Devices
Sep 11, 2019



Abstract:Automated in-vitro cell detection and counting have been a key theme for artificial and intelligent biological analysis such as biopsy, drug analysis and decease diagnosis. Along with the rapid development of microfluidics and lab-on-chip technologies, in-vitro live cell analysis has been one of the critical tasks for both research and industry communities. However, it is a great challenge to obtain and then predict the precise information of live cells from numerous microscopic videos and images. In this paper, we investigated in-vitro detection of white blood cells using deep neural networks, and discussed how state-of-the-art machine learning techniques could fulfil the needs of medical diagnosis. The approach we used in this study was based on Faster Region-based Convolutional Neural Networks (Faster RCNNs), and a transfer learning process was applied to apply this technique to the microscopic detection of blood cells. Our experimental results demonstrated that fast and efficient analysis of blood cells via automated microscopic imaging can achieve much better accuracy and faster speed than the conventionally applied methods, implying a promising future of this technology to be applied to the microfluidic point-of-care medical devices.
Computer-Aided Automated Detection of Gene-Controlled Social Actions of Drosophila
Sep 11, 2019

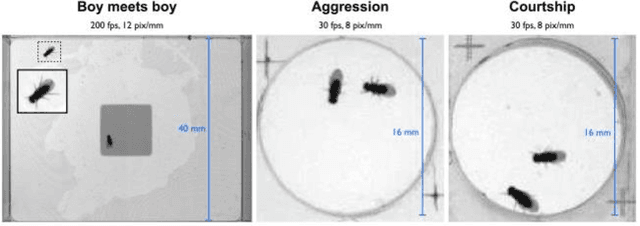
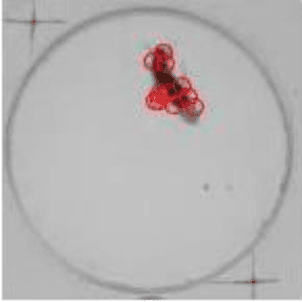
Abstract:Gene expression of social actions in Drosophilae has been attracting wide interest from biologists, medical scientists and psychologists. Gene-edited Drosophilae have been used as a test platform for experimental investigation. For example, Parkinson's genes can be embedded into a group of newly bred Drosophilae for research purpose. However, human observation of numerous tiny Drosophilae for a long term is an arduous work, and the dependence on human's acute perception is highly unreliable. As a result, an automated system of social action detection using machine learning has been highly demanded. In this study, we propose to automate the detection and classification of two innate aggressive actions demonstrated by Drosophilae. Robust keypoint detection is achieved using selective spatio-temporal interest points (sSTIP) which are then described using the 3D Scale Invariant Feature Transform (3D-SIFT) descriptors. Dimensionality reduction is performed using Spectral Regression Kernel Discriminant Analysis (SR-KDA) and classification is done using the nearest centre rule. The classification accuracy shown demonstrates the feasibility of the proposed system.
* published on International Conference on Smart Cities at Cambridge 2018
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge