Tiago Alves
EZClone: Improving DNN Model Extraction Attack via Shape Distillation from GPU Execution Profiles
Apr 06, 2023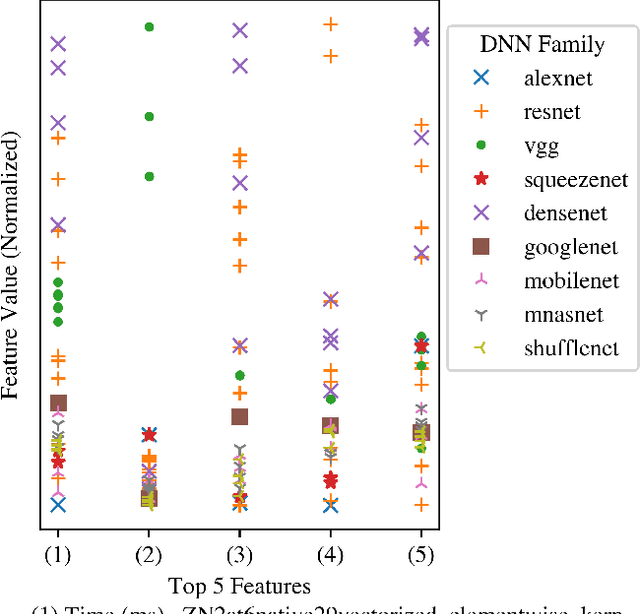
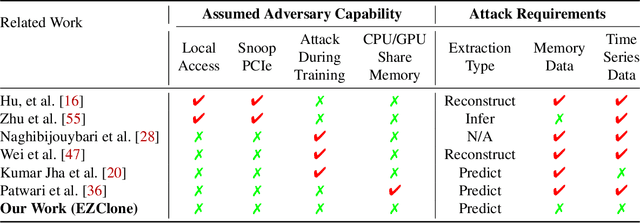

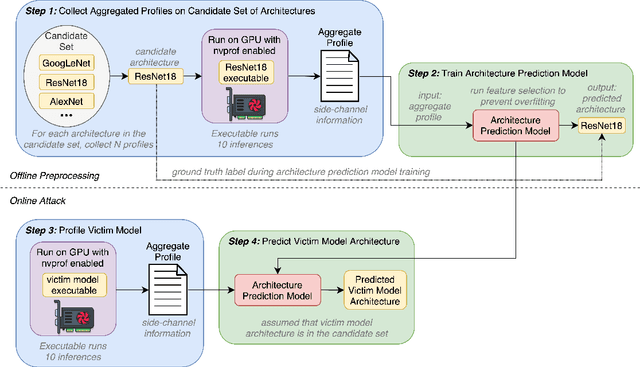
Abstract:Deep Neural Networks (DNNs) have become ubiquitous due to their performance on prediction and classification problems. However, they face a variety of threats as their usage spreads. Model extraction attacks, which steal DNNs, endanger intellectual property, data privacy, and security. Previous research has shown that system-level side-channels can be used to leak the architecture of a victim DNN, exacerbating these risks. We propose two DNN architecture extraction techniques catering to various threat models. The first technique uses a malicious, dynamically linked version of PyTorch to expose a victim DNN architecture through the PyTorch profiler. The second, called EZClone, exploits aggregate (rather than time-series) GPU profiles as a side-channel to predict DNN architecture, employing a simple approach and assuming little adversary capability as compared to previous work. We investigate the effectiveness of EZClone when minimizing the complexity of the attack, when applied to pruned models, and when applied across GPUs. We find that EZClone correctly predicts DNN architectures for the entire set of PyTorch vision architectures with 100% accuracy. No other work has shown this degree of architecture prediction accuracy with the same adversarial constraints or using aggregate side-channel information. Prior work has shown that, once a DNN has been successfully cloned, further attacks such as model evasion or model inversion can be accelerated significantly.
Hardening DNNs against Transfer Attacks during Network Compression using Greedy Adversarial Pruning
Jun 15, 2022

Abstract:The prevalence and success of Deep Neural Network (DNN) applications in recent years have motivated research on DNN compression, such as pruning and quantization. These techniques accelerate model inference, reduce power consumption, and reduce the size and complexity of the hardware necessary to run DNNs, all with little to no loss in accuracy. However, since DNNs are vulnerable to adversarial inputs, it is important to consider the relationship between compression and adversarial robustness. In this work, we investigate the adversarial robustness of models produced by several irregular pruning schemes and by 8-bit quantization. Additionally, while conventional pruning removes the least important parameters in a DNN, we investigate the effect of an unconventional pruning method: removing the most important model parameters based on the gradient on adversarial inputs. We call this method Greedy Adversarial Pruning (GAP) and we find that this pruning method results in models that are resistant to transfer attacks from their uncompressed counterparts.
Dynamic Prediction of ICU Mortality Risk Using Domain Adaptation
Dec 20, 2019



Abstract:Early recognition of risky trajectories during an Intensive Care Unit (ICU) stay is one of the key steps towards improving patient survival. Learning trajectories from physiological signals continuously measured during an ICU stay requires learning time-series features that are robust and discriminative across diverse patient populations. Patients within different ICU populations (referred here as domains) vary by age, conditions and interventions. Thus, mortality prediction models using patient data from a particular ICU population may perform suboptimally in other populations because the features used to train such models have different distributions across the groups. In this paper, we explore domain adaptation strategies in order to learn mortality prediction models that extract and transfer complex temporal features from multivariate time-series ICU data. Features are extracted in a way that the state of the patient in a certain time depends on the previous state. This enables dynamic predictions and creates a mortality risk space that describes the risk of a patient at a particular time. Experiments based on cross-ICU populations reveals that our model outperforms all considered baselines. Gains in terms of AUC range from 4% to 8% for early predictions when compared with a recent state-of-the-art representative for ICU mortality prediction. In particular, models for the Cardiac ICU population achieve AUC numbers as high as 0.88, showing excellent clinical utility for early mortality prediction. Finally, we present an explanation of factors contributing to the possible ICU outcomes, so that our models can be used to complement clinical reasoning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge