Thomas Wolfgruber
Deep Learning Predicts Mammographic Breast Density in Clinical Breast Ultrasound Images
Oct 31, 2024



Abstract:Background: Mammographic breast density, as defined by the American College of Radiology's Breast Imaging Reporting and Data System (BI-RADS), is one of the strongest risk factors for breast cancer, but is derived from mammographic images. Breast ultrasound (BUS) is an alternative breast cancer screening modality, particularly useful for early detection in low-resource, rural contexts. The purpose of this study was to explore an artificial intelligence (AI) model to predict BI-RADS mammographic breast density category from clinical, handheld BUS imaging. Methods: All data are sourced from the Hawaii and Pacific Islands Mammography Registry. We compared deep learning methods from BUS imaging, as well as machine learning models from image statistics alone. The use of AI-derived BUS density as a risk factor for breast cancer was then compared to clinical BI-RADS breast density while adjusting for age. The BUS data were split by individual into 70/20/10% groups for training, validation, and testing. Results: 405,120 clinical BUS images from 14.066 women were selected for inclusion in this study, resulting in 9.846 women for training (302,574 images), 2,813 for validation (11,223 images), and 1,406 for testing (4,042 images). On the held-out testing set, the strongest AI model achieves AUROC 0.854 predicting BI-RADS mammographic breast density from BUS imaging and outperforms all shallow machine learning methods based on image statistics. In cancer risk prediction, age-adjusted AI BUS breast density predicted 5-year breast cancer risk with 0.633 AUROC, as compared to 0.637 AUROC from age-adjusted clinical breast density. Conclusions: BI-RADS mammographic breast density can be estimated from BUS imaging with high accuracy using a deep learning model. Furthermore, we demonstrate that AI-derived BUS breast density is predictive of 5-year breast cancer risk in our population.
Learning a Clinically-Relevant Concept Bottleneck for Lesion Detection in Breast Ultrasound
Jun 29, 2024Abstract:Detecting and classifying lesions in breast ultrasound images is a promising application of artificial intelligence (AI) for reducing the burden of cancer in regions with limited access to mammography. Such AI systems are more likely to be useful in a clinical setting if their predictions can be explained to a radiologist. This work proposes an explainable AI model that provides interpretable predictions using a standard lexicon from the American College of Radiology's Breast Imaging and Reporting Data System (BI-RADS). The model is a deep neural network featuring a concept bottleneck layer in which known BI-RADS features are predicted before making a final cancer classification. This enables radiologists to easily review the predictions of the AI system and potentially fix errors in real time by modifying the concept predictions. In experiments, a model is developed on 8,854 images from 994 women with expert annotations and histological cancer labels. The model outperforms state-of-the-art lesion detection frameworks with 48.9 average precision on the held-out testing set, and for cancer classification, concept intervention is shown to increase performance from 0.876 to 0.885 area under the receiver operating characteristic curve. Training and evaluation code is available at https://github.com/hawaii-ai/bus-cbm.
Quantitative Imaging Principles Improves Medical Image Learning
Jun 14, 2022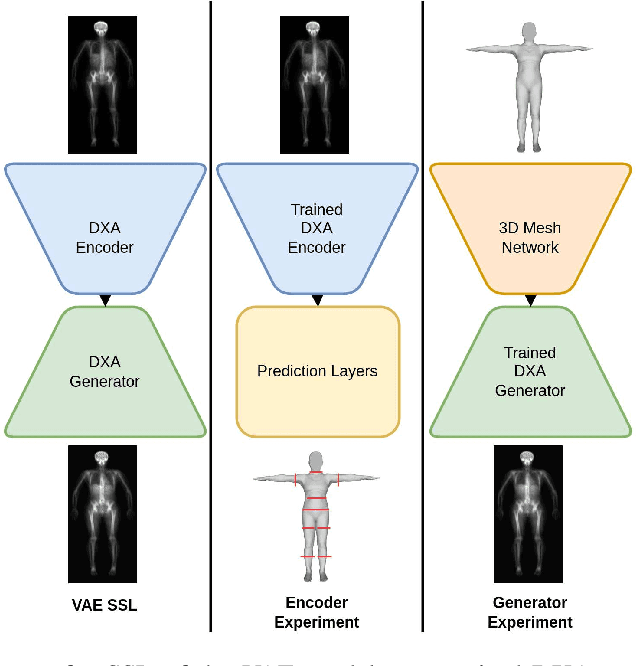
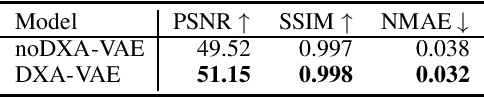
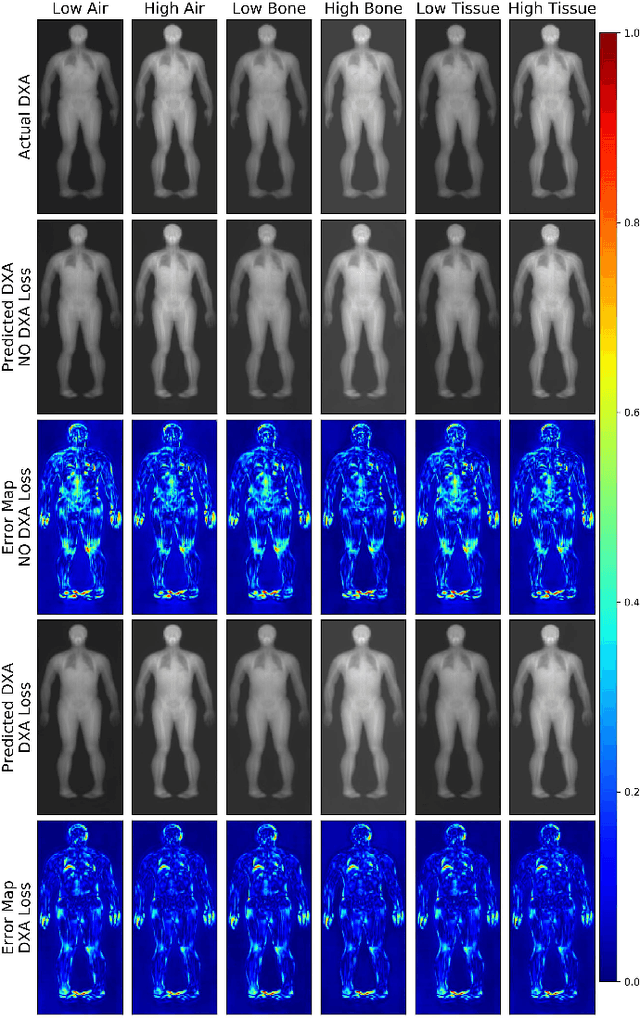
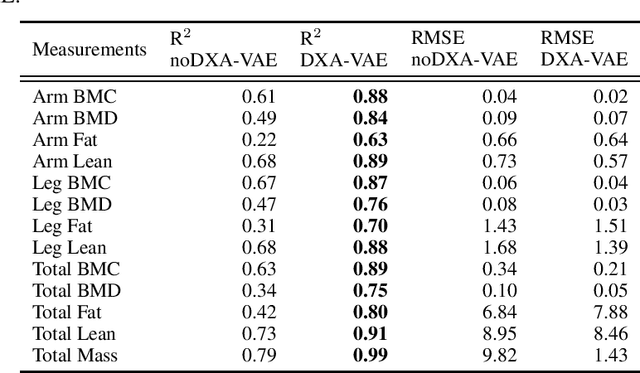
Abstract:Fundamental differences between natural and medical images have recently favored the use of self-supervised learning (SSL) over ImageNet transfer learning for medical image applications. Differences between image types are primarily due to the imaging modality and medical images utilize a wide range of physics based techniques while natural images are captured using only visible light. While many have demonstrated that SSL on medical images has resulted in better downstream task performance, our work suggests that more performance can be gained. The scientific principles which are used to acquire medical images are not often considered when constructing learning problems. For this reason, we propose incorporating quantitative imaging principles during generative SSL to improve image quality and quantitative biological accuracy. We show that this training schema results in better starting states for downstream supervised training on limited data. Our model also generates images that validate on clinical quantitative analysis software.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge