Thanh Ma
REBot: From RAG to CatRAG with Semantic Enrichment and Graph Routing
Oct 02, 2025Abstract:Academic regulation advising is essential for helping students interpret and comply with institutional policies, yet building effective systems requires domain specific regulatory resources. To address this challenge, we propose REBot, an LLM enhanced advisory chatbot powered by CatRAG, a hybrid retrieval reasoning framework that integrates retrieval augmented generation with graph based reasoning. CatRAG unifies dense retrieval and graph reasoning, supported by a hierarchical, category labeled knowledge graph enriched with semantic features for domain alignment. A lightweight intent classifier routes queries to the appropriate retrieval modules, ensuring both factual accuracy and contextual depth. We construct a regulation specific dataset and evaluate REBot on classification and question answering tasks, achieving state of the art performance with an F1 score of 98.89%. Finally, we implement a web application that demonstrates the practical value of REBot in real world academic advising scenarios.
BOLIMES: Boruta and LIME optiMized fEature Selection for Gene Expression Classification
Feb 18, 2025Abstract:Gene expression classification is a pivotal yet challenging task in bioinformatics, primarily due to the high dimensionality of genomic data and the risk of overfitting. To bridge this gap, we propose BOLIMES, a novel feature selection algorithm designed to enhance gene expression classification by systematically refining the feature subset. Unlike conventional methods that rely solely on statistical ranking or classifier-specific selection, we integrate the robustness of Boruta with the interpretability of LIME, ensuring that only the most relevant and influential genes are retained. BOLIMES first employs Boruta to filter out non-informative genes by comparing each feature against its randomized counterpart, thus preserving valuable information. It then uses LIME to rank the remaining genes based on their local importance to the classifier. Finally, an iterative classification evaluation determines the optimal feature subset by selecting the number of genes that maximizes predictive accuracy. By combining exhaustive feature selection with interpretability-driven refinement, our solution effectively balances dimensionality reduction with high classification performance, offering a powerful solution for high-dimensional gene expression analysis.
Region-Based Merging of Open-Domain Terminological Knowledge
May 06, 2022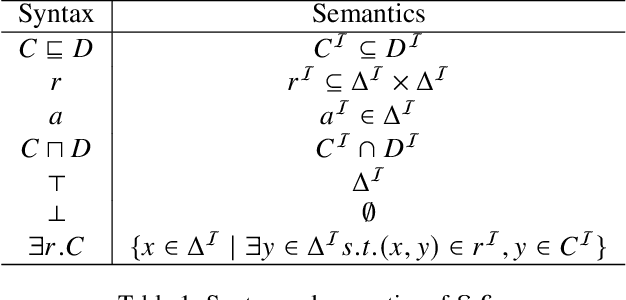
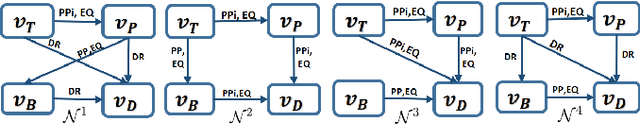
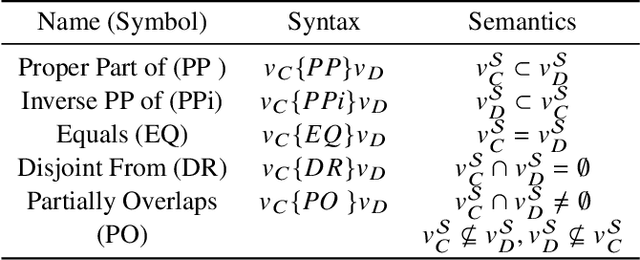
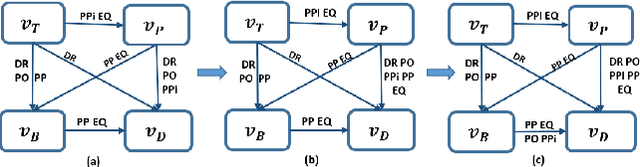
Abstract:This paper introduces a novel method for merging open-domain terminological knowledge. It takes advantage of the Region Connection Calculus (RCC5), a formalism used to represent regions in a topological space and to reason about their set-theoretic relationships. To this end, we first propose a faithful translation of terminological knowledge provided by several and potentially conflicting sources into region spaces. The merging is then performed on these spaces, and the result is translated back into the underlying language of the input sources. Our approach allows us to benefit from the expressivity and the flexibility of RCC5 while dealing with conflicting knowledge in a principled way.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge