Taiyao Wang
Predicting Antimicrobial Resistance in the Intensive Care Unit
Nov 05, 2021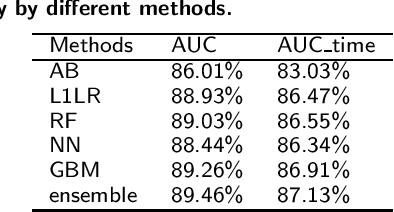
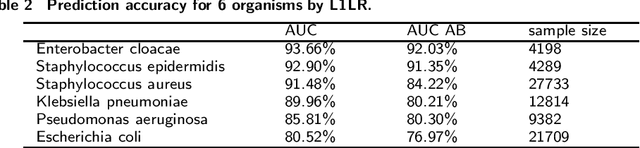

Abstract:Antimicrobial resistance (AMR) is a risk for patients and a burden for the healthcare system. However, AMR assays typically take several days. This study develops predictive models for AMR based on easily available clinical and microbiological predictors, including patient demographics, hospital stay data, diagnoses, clinical features, and microbiological/antimicrobial characteristics and compares those models to a naive antibiogram based model using only microbiological/antimicrobial characteristics. The ability to predict the resistance accurately prior to culturing could inform clinical decision-making and shorten time to action. The machine learning algorithms employed here show improved classification performance (area under the receiver operating characteristic curve 0.88-0.89) versus the naive model (area under the receiver operating characteristic curve 0.86) for 6 organisms and 10 antibiotics using the Philips eICU Research Institute (eRI) database. This method can help guide antimicrobial treatment, with the objective of improving patient outcomes and reducing the usage of unnecessary or ineffective antibiotics.
Convergence of Parameter Estimates for Regularized Mixed Linear Regression Models
Mar 21, 2019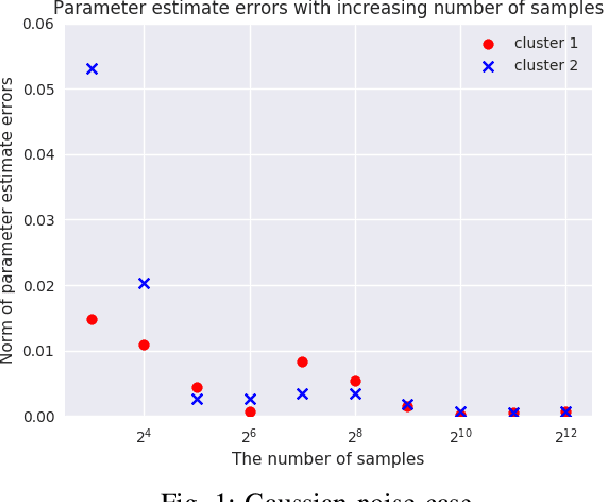
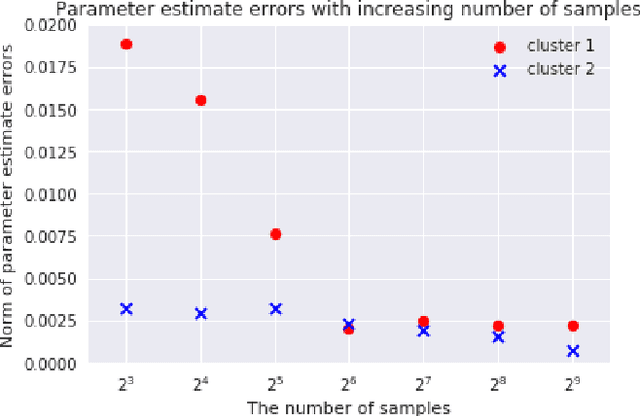
Abstract:We consider {\em Mixed Linear Regression (MLR)}, where training data have been generated from a mixture of distinct linear models (or clusters) and we seek to identify the corresponding coefficient vectors. We introduce a {\em Mixed Integer Programming (MIP)} formulation for MLR subject to regularization constraints on the coefficient vectors. We establish that as the number of training samples grows large, the MIP solution converges to the true coefficient vectors in the absence of noise. Subject to slightly stronger assumptions, we also establish that the MIP identifies the clusters from which the training samples were generated. In the special case where training data come from a single cluster, we establish that the corresponding MIP yields a solution that converges to the true coefficient vector even when training data are perturbed by (martingale difference) noise. We provide a counterexample indicating that in the presence of noise, the MIP may fail to produce the true coefficient vectors for more than one clusters. We also provide numerical results testing the MIP solutions in synthetic examples with noise.
Prescriptive Cluster-Dependent Support Vector Machines with an Application to Reducing Hospital Readmissions
Mar 21, 2019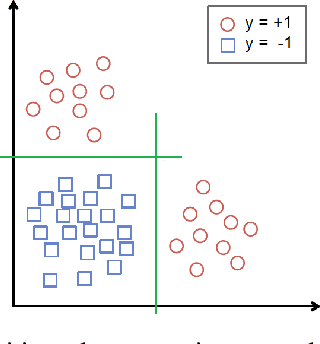
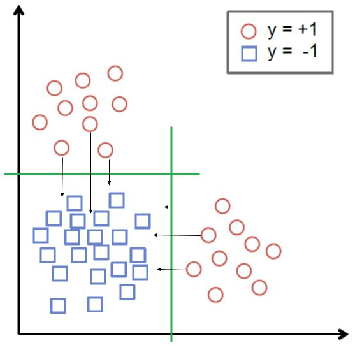
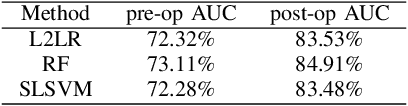
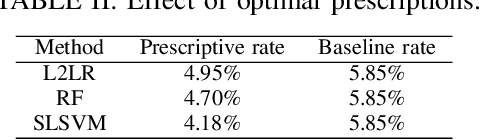
Abstract:We augment linear Support Vector Machine (SVM) classifiers by adding three important features: (i) we introduce a regularization constraint to induce a sparse classifier; (ii) we devise a method that partitions the positive class into clusters and selects a sparse SVM classifier for each cluster; and (iii) we develop a method to optimize the values of controllable variables in order to reduce the number of data points which are predicted to have an undesirable outcome, which, in our setting, coincides with being in the positive class. The latter feature leads to personalized prescriptions/recommendations. We apply our methods to the problem of predicting and preventing hospital readmissions within 30-days from discharge for patients that underwent a general surgical procedure. To that end, we leverage a large dataset containing over 2.28 million patients who had surgeries in the period 2011--2014 in the U.S. The dataset has been collected as part of the American College of Surgeons National Surgical Quality Improvement Program (NSQIP).
Predicting Chronic Disease Hospitalizations from Electronic Health Records: An Interpretable Classification Approach
Jan 03, 2018
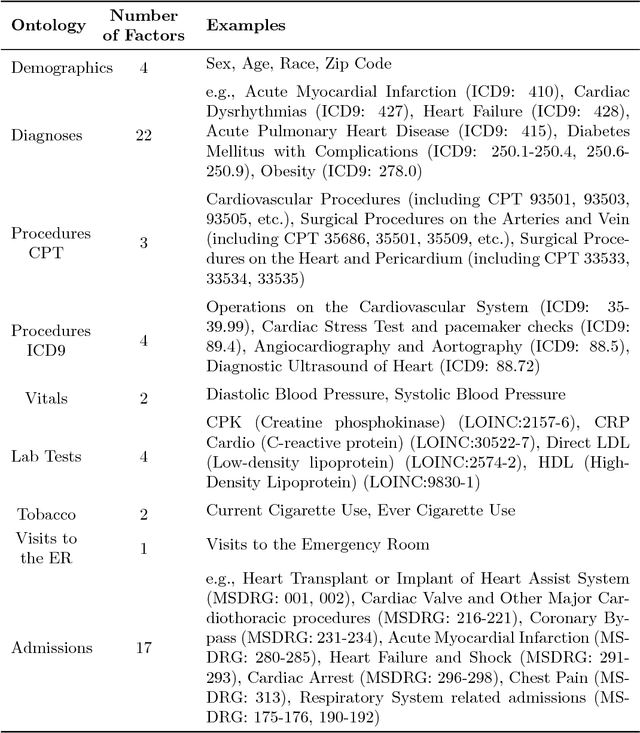
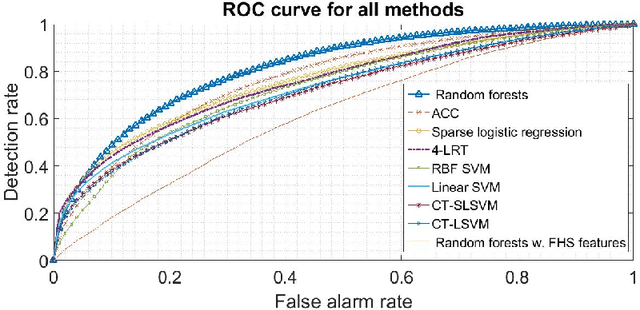
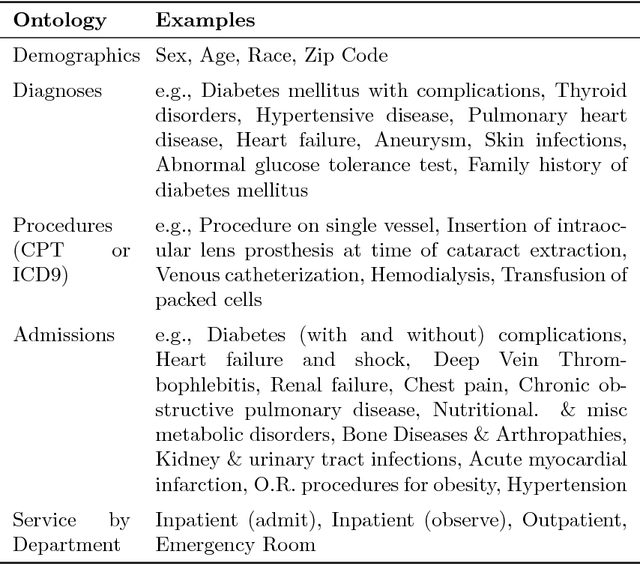
Abstract:Urban living in modern large cities has significant adverse effects on health, increasing the risk of several chronic diseases. We focus on the two leading clusters of chronic disease, heart disease and diabetes, and develop data-driven methods to predict hospitalizations due to these conditions. We base these predictions on the patients' medical history, recent and more distant, as described in their Electronic Health Records (EHR). We formulate the prediction problem as a binary classification problem and consider a variety of machine learning methods, including kernelized and sparse Support Vector Machines (SVM), sparse logistic regression, and random forests. To strike a balance between accuracy and interpretability of the prediction, which is important in a medical setting, we propose two novel methods: K-LRT, a likelihood ratio test-based method, and a Joint Clustering and Classification (JCC) method which identifies hidden patient clusters and adapts classifiers to each cluster. We develop theoretical out-of-sample guarantees for the latter method. We validate our algorithms on large datasets from the Boston Medical Center, the largest safety-net hospital system in New England.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge