Sydney Anuyah
Benchmarking LLMs for Pairwise Causal Discovery in Biomedical and Multi-Domain Contexts
Jan 21, 2026Abstract:The safe deployment of large language models (LLMs) in high-stakes fields like biomedicine, requires them to be able to reason about cause and effect. We investigate this ability by testing 13 open-source LLMs on a fundamental task: pairwise causal discovery (PCD) from text. Our benchmark, using 12 diverse datasets, evaluates two core skills: 1) \textbf{Causal Detection} (identifying if a text contains a causal link) and 2) \textbf{Causal Extraction} (pulling out the exact cause and effect phrases). We tested various prompting methods, from simple instructions (zero-shot) to more complex strategies like Chain-of-Thought (CoT) and Few-shot In-Context Learning (FICL). The results show major deficiencies in current models. The best model for detection, DeepSeek-R1-Distill-Llama-70B, only achieved a mean score of 49.57\% ($C_{detect}$), while the best for extraction, Qwen2.5-Coder-32B-Instruct, reached just 47.12\% ($C_{extract}$). Models performed best on simple, explicit, single-sentence relations. However, performance plummeted for more difficult (and realistic) cases, such as implicit relationships, links spanning multiple sentences, and texts containing multiple causal pairs. We provide a unified evaluation framework, built on a dataset validated with high inter-annotator agreement ($κ\ge 0.758$), and make all our data, code, and prompts publicly available to spur further research. \href{https://github.com/sydneyanuyah/CausalDiscovery}{Code available here: https://github.com/sydneyanuyah/CausalDiscovery}
Domain-Specific Knowledge Graphs in RAG-Enhanced Healthcare LLMs
Jan 21, 2026Abstract:Large Language Models (LLMs) generate fluent answers but can struggle with trustworthy, domain-specific reasoning. We evaluate whether domain knowledge graphs (KGs) improve Retrieval-Augmented Generation (RAG) for healthcare by constructing three PubMed-derived graphs: $\mathbb{G}_1$ (T2DM), $\mathbb{G}_2$ (Alzheimer's disease), and $\mathbb{G}_3$ (AD+T2DM). We design two probes: Probe 1 targets merged AD T2DM knowledge, while Probe 2 targets the intersection of $\mathbb{G}_1$ and $\mathbb{G}_2$. Seven instruction-tuned LLMs are tested across retrieval sources {No-RAG, $\mathbb{G}_1$, $\mathbb{G}_2$, $\mathbb{G}_1$ + $\mathbb{G}_2$, $\mathbb{G}_3$, $\mathbb{G}_1$+$\mathbb{G}_2$ + $\mathbb{G}_3$} and three decoding temperatures. Results show that scope alignment between probe and KG is decisive: precise, scope-matched retrieval (notably $\mathbb{G}_2$) yields the most consistent gains, whereas indiscriminate graph unions often introduce distractors that reduce accuracy. Larger models frequently match or exceed KG-RAG with a No-RAG baseline on Probe 1, indicating strong parametric priors, whereas smaller/mid-sized models benefit more from well-scoped retrieval. Temperature plays a secondary role; higher values rarely help. We conclude that precision-first, scope-matched KG-RAG is preferable to breadth-first unions, and we outline practical guidelines for graph selection, model sizing, and retrieval/reranking. Code and Data available here - https://github.com/sydneyanuyah/RAGComparison
An Empirical Study of Causal Relation Extraction Transfer: Design and Data
Mar 08, 2025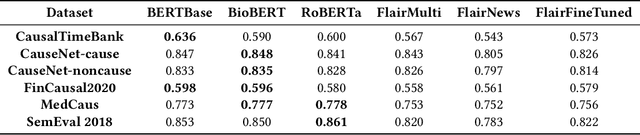
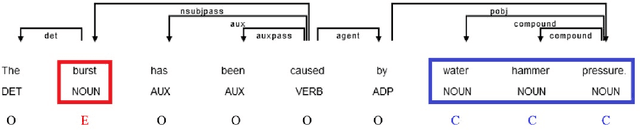
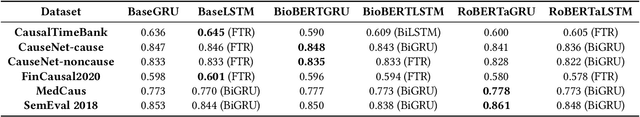
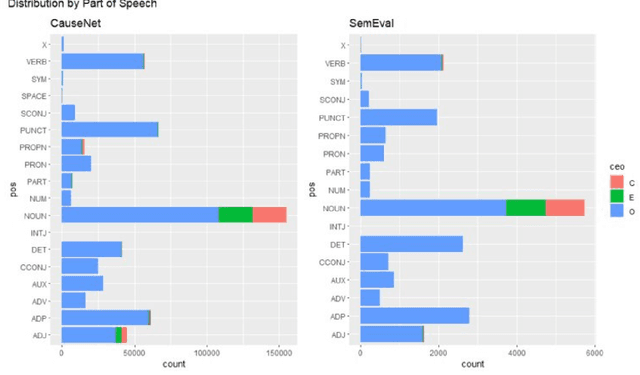
Abstract:We conduct an empirical analysis of neural network architectures and data transfer strategies for causal relation extraction. By conducting experiments with various contextual embedding layers and architectural components, we show that a relatively straightforward BioBERT-BiGRU relation extraction model generalizes better than other architectures across varying web-based sources and annotation strategies. Furthermore, we introduce a metric for evaluating transfer performance, $F1_{phrase}$ that emphasizes noun phrase localization rather than directly matching target tags. Using this metric, we can conduct data transfer experiments, ultimately revealing that augmentation with data with varying domains and annotation styles can improve performance. Data augmentation is especially beneficial when an adequate proportion of implicitly and explicitly causal sentences are included.
Advancing clinical trial outcomes using deep learning and predictive modelling: bridging precision medicine and patient-centered care
Dec 09, 2024Abstract:The integration of artificial intelligence [AI] into clinical trials has revolutionized the process of drug development and personalized medicine. Among these advancements, deep learning and predictive modelling have emerged as transformative tools for optimizing clinical trial design, patient recruitment, and real-time monitoring. This study explores the application of deep learning techniques, such as convolutional neural networks [CNNs] and transformerbased models, to stratify patients, forecast adverse events, and personalize treatment plans. Furthermore, predictive modelling approaches, including survival analysis and time-series forecasting, are employed to predict trial outcomes, enhancing efficiency and reducing trial failure rates. To address challenges in analysing unstructured clinical data, such as patient notes and trial protocols, natural language processing [NLP] techniques are utilized for extracting actionable insights. A custom dataset comprising structured patient demographics, genomic data, and unstructured text is curated for training and validating these models. Key metrics, including precision, recall, and F1 scores, are used to evaluate model performance, while trade-offs between accuracy and computational efficiency are examined to identify the optimal model for clinical deployment. This research underscores the potential of AI-driven methods to streamline clinical trial workflows, improve patient-centric outcomes, and reduce costs associated with trial inefficiencies. The findings provide a robust framework for integrating predictive analytics into precision medicine, paving the way for more adaptive and efficient clinical trials. By bridging the gap between technological innovation and real-world applications, this study contributes to advancing the role of AI in healthcare, particularly in fostering personalized care and improving overall trial success rates.
* 22 pages excluding references, 11 figures, 6 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge