Susan McKeever
What Makes a Meme a Meme? Identifying Memes for Memetics-Aware Dataset Creation
Jul 16, 2024Abstract:Warning: This paper contains memes that may be offensive to some readers. Multimodal Internet Memes are now a ubiquitous fixture in online discourse. One strand of meme-based research is the classification of memes according to various affects, such as sentiment and hate, supported by manually compiled meme datasets. Understanding the unique characteristics of memes is crucial for meme classification. Unlike other user-generated content, memes spread via memetics, i.e. the process by which memes are imitated and transformed into symbols used to create new memes. In effect, there exists an ever-evolving pool of visual and linguistic symbols that underpin meme culture and are crucial to interpreting the meaning of individual memes. The current approach of training supervised learning models on static datasets, without taking memetics into account, limits the depth and accuracy of meme interpretation. We argue that meme datasets must contain genuine memes, as defined via memetics, so that effective meme classifiers can be built. In this work, we develop a meme identification protocol which distinguishes meme from non-memetic content by recognising the memetics within it. We apply our protocol to random samplings of the leading 7 meme classification datasets and observe that more than half (50. 4\%) of the evaluated samples were found to contain no signs of memetics. Our work also provides a meme typology grounded in memetics, providing the basis for more effective approaches to the interpretation of memes and the creation of meme datasets.
Unimodal Intermediate Training for Multimodal Meme Sentiment Classification
Aug 01, 2023



Abstract:Internet Memes remain a challenging form of user-generated content for automated sentiment classification. The availability of labelled memes is a barrier to developing sentiment classifiers of multimodal memes. To address the shortage of labelled memes, we propose to supplement the training of a multimodal meme classifier with unimodal (image-only and text-only) data. In this work, we present a novel variant of supervised intermediate training that uses relatively abundant sentiment-labelled unimodal data. Our results show a statistically significant performance improvement from the incorporation of unimodal text data. Furthermore, we show that the training set of labelled memes can be reduced by 40% without reducing the performance of the downstream model.
Meme Sentiment Analysis Enhanced with Multimodal Spatial Encoding and Facial Embedding
Mar 03, 2023Abstract:Internet memes are characterised by the interspersing of text amongst visual elements. State-of-the-art multimodal meme classifiers do not account for the relative positions of these elements across the two modalities, despite the latent meaning associated with where text and visual elements are placed. Against two meme sentiment classification datasets, we systematically show performance gains from incorporating the spatial position of visual objects, faces, and text clusters extracted from memes. In addition, we also present facial embedding as an impactful enhancement to image representation in a multimodal meme classifier. Finally, we show that incorporating this spatial information allows our fully automated approaches to outperform their corresponding baselines that rely on additional human validation of OCR-extracted text.
* Published as chapter in ISBN:978-3-031-26438-2
Exploring the Impact of Noise and Degradations on Heart Sound Classification Models
Nov 14, 2022



Abstract:The development of data-driven heart sound classification models has been an active area of research in recent years. To develop such data-driven models in the first place, heart sound signals need to be captured using a signal acquisition device. However, it is almost impossible to capture noise-free heart sound signals due to the presence of internal and external noises in most situations. Such noises and degradations in heart sound signals can potentially reduce the accuracy of data-driven classification models. Although different techniques have been proposed in the literature to address the noise issue, how and to what extent different noise and degradations in heart sound signals impact the accuracy of data-driven classification models remains unexplored. To answer this question, we produced a synthetic heart sound dataset including normal and abnormal heart sounds contaminated with a large variety of noise and degradations. We used this dataset to investigate the impact of noise and degradation in heart sound recordings on the performance of different classification models. The results show different noises and degradations affect the performance of heart sound classification models to a different extent; some are more problematic for classification models, and others are less destructive. Comparing the findings of this study with the results of a survey we previously carried out with a group of clinicians shows noise and degradations that are more detrimental to classification models are also more disruptive to accurate auscultation. The findings of this study can be leveraged to develop targeted heart sound quality enhancement approaches - which adapt the type and aggressiveness of quality enhancement based on the characteristics of noise and degradation in heart sound signals.
Wider Vision: Enriching Convolutional Neural Networks via Alignment to External Knowledge Bases
Feb 22, 2021
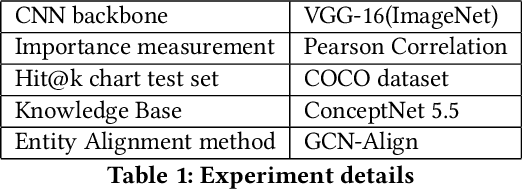


Abstract:Deep learning models suffer from opaqueness. For Convolutional Neural Networks (CNNs), current research strategies for explaining models focus on the target classes within the associated training dataset. As a result, the understanding of hidden feature map activations is limited by the discriminative knowledge gleaned during training. The aim of our work is to explain and expand CNNs models via the mirroring or alignment of CNN to an external knowledge base. This will allow us to give a semantic context or label for each visual feature. We can match CNN feature activations to nodes in our external knowledge base. This supports knowledge-based interpretation of the features associated with model decisions. To demonstrate our approach, we build two separate graphs. We use an entity alignment method to align the feature nodes in a CNN with the nodes in a ConceptNet based knowledge graph. We then measure the proximity of CNN graph nodes to semantically meaningful knowledge base nodes. Our results show that in the aligned embedding space, nodes from the knowledge graph are close to the CNN feature nodes that have similar meanings, indicating that nodes from an external knowledge base can act as explanatory semantic references for features in the model. We analyse a variety of graph building methods in order to improve the results from our embedding space. We further demonstrate that by using hierarchical relationships from our external knowledge base, we can locate new unseen classes outside the CNN training set in our embeddings space, based on visual feature activations. This suggests that we can adapt our approach to identify unseen classes based on CNN feature activations. Our demonstrated approach of aligning a CNN with an external knowledge base paves the way to reason about and beyond the trained model, with future adaptations to explainable models and zero-shot learning.
Feature Fusion of Raman Chemical Imaging and Digital Histopathology using Machine Learning for Prostate Cancer Detection
Jan 18, 2021



Abstract:The diagnosis of prostate cancer is challenging due to the heterogeneity of its presentations, leading to the over diagnosis and treatment of non-clinically important disease. Accurate diagnosis can directly benefit a patient's quality of life and prognosis. Towards addressing this issue, we present a learning model for the automatic identification of prostate cancer. While many prostate cancer studies have adopted Raman spectroscopy approaches, none have utilised the combination of Raman Chemical Imaging (RCI) and other imaging modalities. This study uses multimodal images formed from stained Digital Histopathology (DP) and unstained RCI. The approach was developed and tested on a set of 178 clinical samples from 32 patients, containing a range of non-cancerous, Gleason grade 3 (G3) and grade 4 (G4) tissue microarray samples. For each histological sample, there is a pathologist labelled DP - RCI image pair. The hypothesis tested was whether multimodal image models can outperform single modality baseline models in terms of diagnostic accuracy. Binary non-cancer/cancer models and the more challenging G3/G4 differentiation were investigated. Regarding G3/G4 classification, the multimodal approach achieved a sensitivity of 73.8% and specificity of 88.1% while the baseline DP model showed a sensitivity and specificity of 54.1% and 84.7% respectively. The multimodal approach demonstrated a statistically significant 12.7% AUC advantage over the baseline with a value of 85.8% compared to 73.1%, also outperforming models based solely on RCI and median Raman spectra. Feature fusion of DP and RCI does not improve the more trivial task of tumour identification but does deliver an observed advantage in G3/G4 discrimination. Building on these promising findings, future work could include the acquisition of larger datasets for enhanced model generalization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge