Suman S. Thapa
Detecting Glaucoma Using 3D Convolutional Neural Network of Raw SD-OCT Optic Nerve Scans
Oct 14, 2019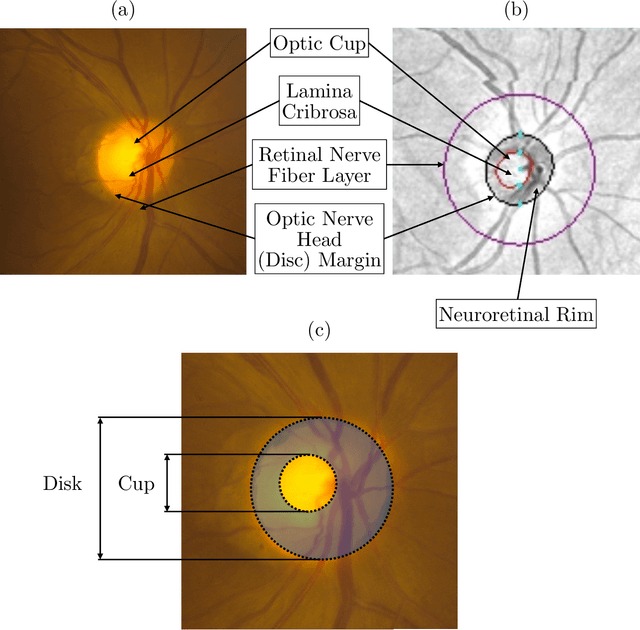
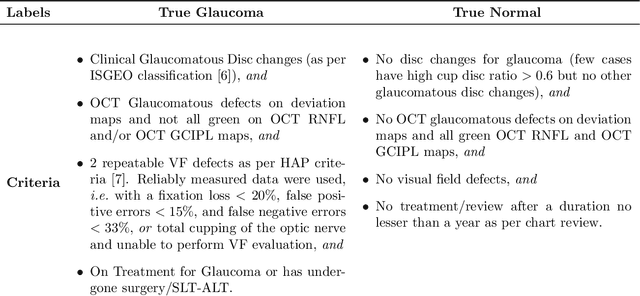
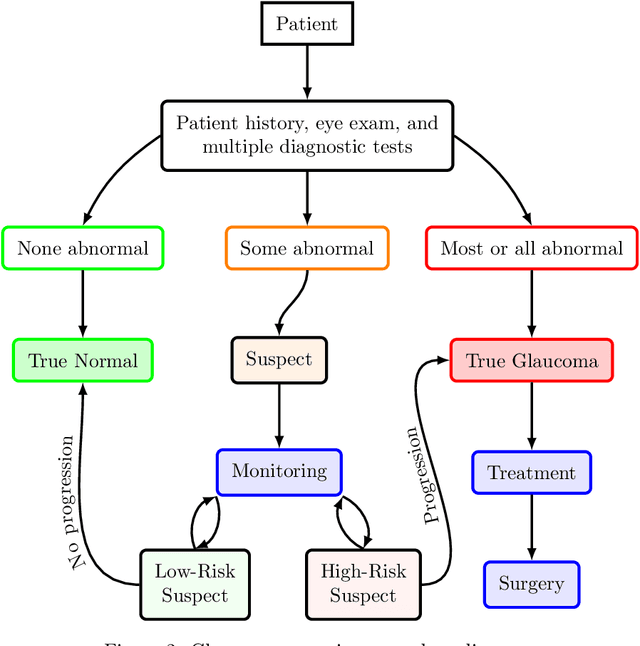
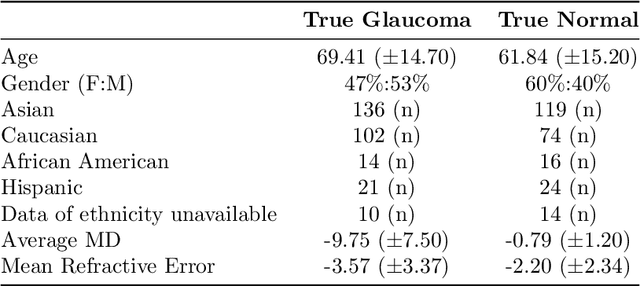
Abstract:We propose developing and validating a three-dimensional (3D) deep learning system using the entire unprocessed OCT optic nerve volumes to distinguish true glaucoma from normals in order to discover any additional imaging biomarkers within the cube through saliency mapping. The algorithm has been validated against 4 additional distinct datasets from different countries using multimodal test results to define glaucoma rather than just the OCT alone. 2076 OCT (Cirrus SD-OCT, Carl Zeiss Meditec, Dublin, CA) cube scans centered over the optic nerve, of 879 eyes (390 healthy and 489 glaucoma) from 487 patients, age 18-84 years, were exported from the Glaucoma Clinic Imaging Database at the Byers Eye Institute, Stanford University, from March 2010 to December 2017. A 3D deep neural network was trained and tested on this unique OCT optic nerve head dataset from Stanford. A total of 3620 scans (all obtained using the Cirrus SD-OCT device) from 1458 eyes obtained from 4 different institutions, from United States (943 scans), Hong Kong (1625 scans), India (672 scans), and Nepal (380 scans) were used for external evaluation. The 3D deep learning system achieved an area under the receiver operation characteristics curve (AUROC) of 0.8883 in the primary Stanford test set identifying true normal from true glaucoma. The system obtained AUROCs of 0.8571, 0.7695, 0.8706, and 0.7965 on OCT cubes from United States, Hong Kong, India, and Nepal, respectively. We also analyzed the performance of the model separately for each myopia severity level as defined by spherical equivalent and the model was able to achieve F1 scores of 0.9673, 0.9491, and 0.8528 on severe, moderate, and mild myopia cases, respectively. Saliency map visualizations highlighted a significant association between the optic nerve lamina cribrosa region in the glaucoma group.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge