Steven Jiang
Multi-Ontology Refined Embeddings (MORE): A Hybrid Multi-Ontology and Corpus-based Semantic Representation for Biomedical Concepts
Apr 14, 2020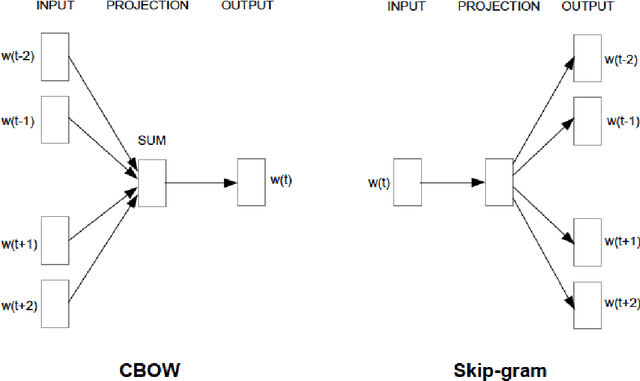
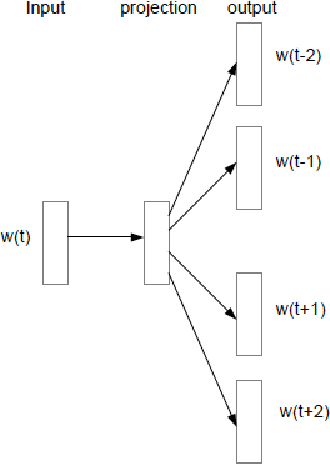
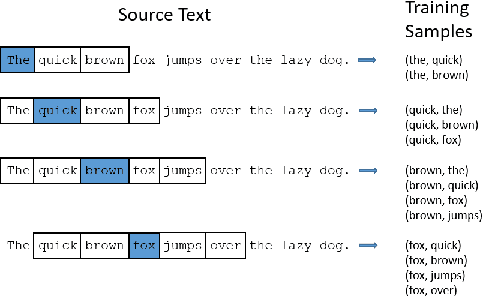
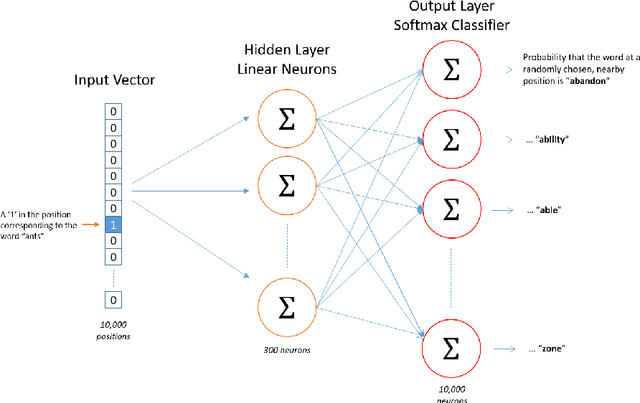
Abstract:Objective: Currently, a major limitation for natural language processing (NLP) analyses in clinical applications is that a concept can be referenced in various forms across different texts. This paper introduces Multi-Ontology Refined Embeddings (MORE), a novel hybrid framework for incorporating domain knowledge from multiple ontologies into a distributional semantic model, learned from a corpus of clinical text. Materials and Methods: We use the RadCore and MIMIC-III free-text datasets for the corpus-based component of MORE. For the ontology-based part, we use the Medical Subject Headings (MeSH) ontology and three state-of-the-art ontology-based similarity measures. In our approach, we propose a new learning objective, modified from the Sigmoid cross-entropy objective function. Results and Discussion: We evaluate the quality of the generated word embeddings using two established datasets of semantic similarities among biomedical concept pairs. On the first dataset with 29 concept pairs, with the similarity scores established by physicians and medical coders, MORE's similarity scores have the highest combined correlation (0.633), which is 5.0% higher than that of the baseline model and 12.4% higher than that of the best ontology-based similarity measure.On the second dataset with 449 concept pairs, MORE's similarity scores have a correlation of 0.481, with the average of four medical residents' similarity ratings, and that outperforms the skip-gram model by 8.1% and the best ontology measure by 6.9%.
Automatic Post-Stroke Lesion Segmentation on MR Images using 3D Residual Convolutional Neural Network
Nov 25, 2019



Abstract:In this paper, we demonstrate the feasibility and performance of deep residual neural networks for volumetric segmentation of irreversibly damaged brain tissue lesions on T1-weighted MRI scans for chronic stroke patients. A total of 239 T1-weighted MRI scans of chronic ischemic stroke patients from a public dataset were retrospectively analyzed by 3D deep convolutional segmentation models with residual learning, using a novel zoom-in&out strategy. Dice similarity coefficient (DSC), Average symmetric surface distance (ASSD), and Hausdorff distance (HD) of the identified lesions were measured by using the manual tracing of lesions as the reference standard. Bootstrapping was employed for all metrics to estimate 95% confidence intervals. The models were assessed on the test set of 31 scans. The average DSC was 0.64 (0.51-0.76) with a median of 0.78. ASSD and HD were 3.6 mm (1.7-6.2 mm) and 20.4 mm (10.0-33.3 mm), respectively. To the best of our knowledge, this performance is the highest achieved on this public dataset. The latest deep learning architecture and techniques were applied for 3D segmentation on MRI scans and demonstrated to be effective for volumetric segmentation of chronic ischemic stroke lesions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge