Stefano Woerner
Navigating Data Scarcity using Foundation Models: A Benchmark of Few-Shot and Zero-Shot Learning Approaches in Medical Imaging
Aug 15, 2024Abstract:Data scarcity is a major limiting factor for applying modern machine learning techniques to clinical tasks. Although sufficient data exists for some well-studied medical tasks, there remains a long tail of clinically relevant tasks with poor data availability. Recently, numerous foundation models have demonstrated high suitability for few-shot learning (FSL) and zero-shot learning (ZSL), potentially making them more accessible to practitioners. However, it remains unclear which foundation model performs best on FSL medical image analysis tasks and what the optimal methods are for learning from limited data. We conducted a comprehensive benchmark study of ZSL and FSL using 16 pretrained foundation models on 19 diverse medical imaging datasets. Our results indicate that BiomedCLIP, a model pretrained exclusively on medical data, performs best on average for very small training set sizes, while very large CLIP models pretrained on LAION-2B perform best with slightly more training samples. However, simply fine-tuning a ResNet-18 pretrained on ImageNet performs similarly with more than five training examples per class. Our findings also highlight the need for further research on foundation models specifically tailored for medical applications and the collection of more datasets to train these models.
Attri-Net: A Globally and Locally Inherently Interpretable Model for Multi-Label Classification Using Class-Specific Counterfactuals
Jun 08, 2024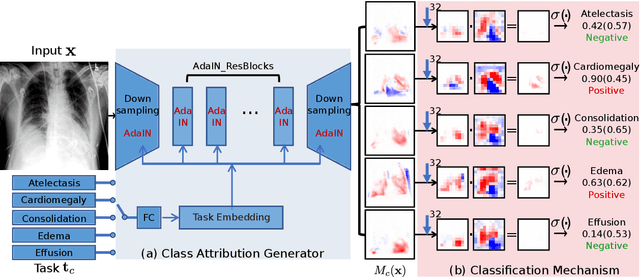
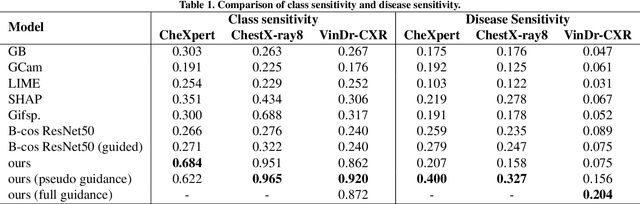
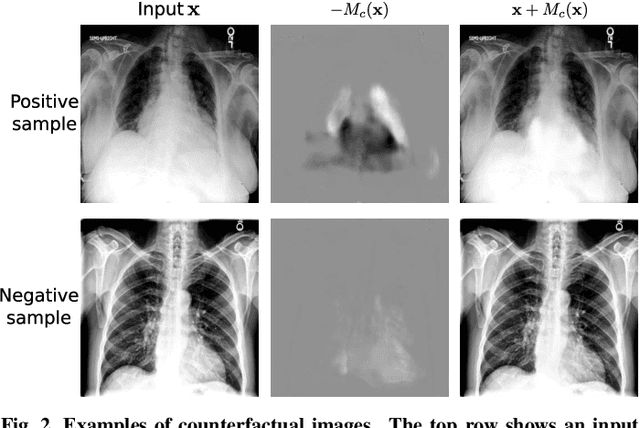

Abstract:Interpretability is crucial for machine learning algorithms in high-stakes medical applications. However, high-performing neural networks typically cannot explain their predictions. Post-hoc explanation methods provide a way to understand neural networks but have been shown to suffer from conceptual problems. Moreover, current research largely focuses on providing local explanations for individual samples rather than global explanations for the model itself. In this paper, we propose Attri-Net, an inherently interpretable model for multi-label classification that provides local and global explanations. Attri-Net first counterfactually generates class-specific attribution maps to highlight the disease evidence, then performs classification with logistic regression classifiers based solely on the attribution maps. Local explanations for each prediction can be obtained by interpreting the attribution maps weighted by the classifiers' weights. Global explanation of whole model can be obtained by jointly considering learned average representations of the attribution maps for each class (called the class centers) and the weights of the linear classifiers. To ensure the model is ``right for the right reason", we further introduce a mechanism to guide the model's explanations to align with human knowledge. Our comprehensive evaluations show that Attri-Net can generate high-quality explanations consistent with clinical knowledge while not sacrificing classification performance.
A comprehensive and easy-to-use multi-domain multi-task medical imaging meta-dataset
Apr 24, 2024Abstract:While the field of medical image analysis has undergone a transformative shift with the integration of machine learning techniques, the main challenge of these techniques is often the scarcity of large, diverse, and well-annotated datasets. Medical images vary in format, size, and other parameters and therefore require extensive preprocessing and standardization, for usage in machine learning. Addressing these challenges, we introduce the Medical Imaging Meta-Dataset (MedIMeta), a novel multi-domain, multi-task meta-dataset. MedIMeta contains 19 medical imaging datasets spanning 10 different domains and encompassing 54 distinct medical tasks, all of which are standardized to the same format and readily usable in PyTorch or other ML frameworks. We perform a technical validation of MedIMeta, demonstrating its utility through fully supervised and cross-domain few-shot learning baselines.
Inherently Interpretable Multi-Label Classification Using Class-Specific Counterfactuals
Mar 01, 2023Abstract:Interpretability is essential for machine learning algorithms in high-stakes application fields such as medical image analysis. However, high-performing black-box neural networks do not provide explanations for their predictions, which can lead to mistrust and suboptimal human-ML collaboration. Post-hoc explanation techniques, which are widely used in practice, have been shown to suffer from severe conceptual problems. Furthermore, as we show in this paper, current explanation techniques do not perform adequately in the multi-label scenario, in which multiple medical findings may co-occur in a single image. We propose Attri-Net, an inherently interpretable model for multi-label classification. Attri-Net is a powerful classifier that provides transparent, trustworthy, and human-understandable explanations. The model first generates class-specific attribution maps based on counterfactuals to identify which image regions correspond to certain medical findings. Then a simple logistic regression classifier is used to make predictions based solely on these attribution maps. We compare Attri-Net to five post-hoc explanation techniques and one inherently interpretable classifier on three chest X-ray datasets. We find that Attri-Net produces high-quality multi-label explanations consistent with clinical knowledge and has comparable classification performance to state-of-the-art classification models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge