Somoballi Ghoshal
Lumbar Spine Tumor Segmentation and Localization in T2 MRI Images Using AI
May 07, 2024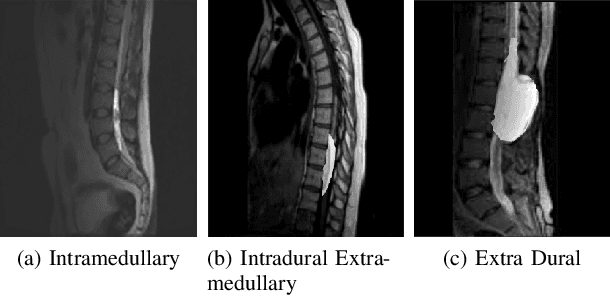

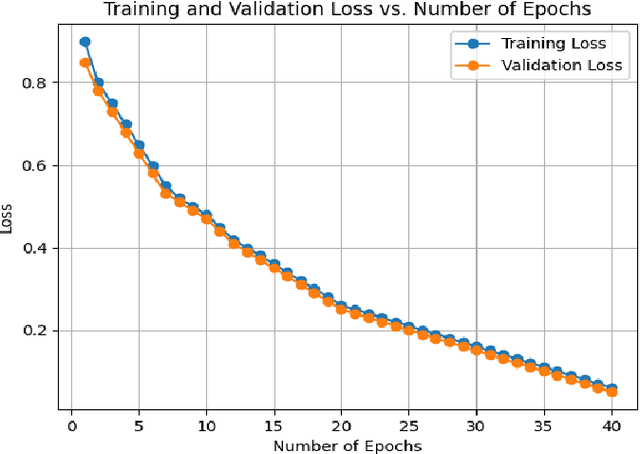

Abstract:In medical imaging, segmentation and localization of spinal tumors in three-dimensional (3D) space pose significant computational challenges, primarily stemming from limited data availability. In response, this study introduces a novel data augmentation technique, aimed at automating spine tumor segmentation and localization through AI approaches. Leveraging a fusion of fuzzy c-means clustering and Random Forest algorithms, the proposed method achieves successful spine tumor segmentation based on predefined masks initially delineated by domain experts in medical imaging. Subsequently, a Convolutional Neural Network (CNN) architecture is employed for tumor classification. Moreover, 3D vertebral segmentation and labeling techniques are used to help pinpoint the exact location of the tumors in the lumbar spine. Results indicate a remarkable performance, with 99% accuracy for tumor segmentation, 98% accuracy for tumor classification, and 99% accuracy for tumor localization achieved with the proposed approach. These metrics surpass the efficacy of existing state-of-the-art techniques, as evidenced by superior Dice Score, Class Accuracy, and Intersection over Union (IOU) on class accuracy metrics. This innovative methodology holds promise for enhancing the diagnostic capabilities in detecting and characterizing spinal tumors, thereby facilitating more effective clinical decision-making.
Panoptic Segmentation and Labelling of Lumbar Spine Vertebrae using Modified Attention Unet
Apr 28, 2024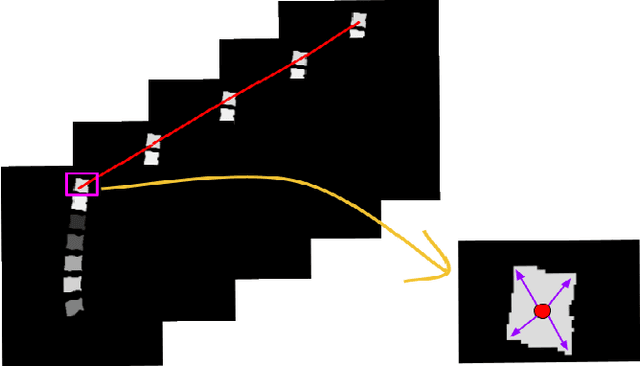


Abstract:Segmentation and labeling of vertebrae in MRI images of the spine are critical for the diagnosis of illnesses and abnormalities. These steps are indispensable as MRI technology provides detailed information about the tissue structure of the spine. Both supervised and unsupervised segmentation methods exist, yet acquiring sufficient data remains challenging for achieving high accuracy. In this study, we propose an enhancing approach based on modified attention U-Net architecture for panoptic segmentation of 3D sliced MRI data of the lumbar spine. Our method achieves an impressive accuracy of 99.5\% by incorporating novel masking logic, thus significantly advancing the state-of-the-art in vertebral segmentation and labeling. This contributes to more precise and reliable diagnosis and treatment planning.
Fast 3D Volumetric Image Reconstruction from 2D MRI Slices by Parallel Processing
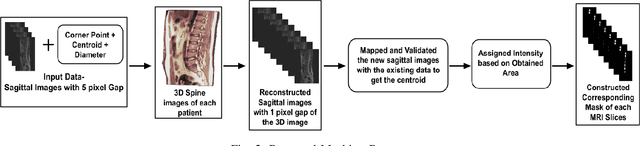
Mar 16, 2023Abstract:Magnetic Resonance Imaging (MRI) is a technology for non-invasive imaging of anatomical features in detail. It can help in functional analysis of organs of a specimen but it is very costly. In this work, methods for (i) virtual three-dimensional (3D) reconstruction from a single sequence of two-dimensional (2D) slices of MR images of a human spine and brain along a single axis, and (ii) generation of missing inter-slice data are proposed. Our approach helps in preserving the edges, shape, size, as well as the internal tissue structures of the object being captured. The sequence of original 2D slices along a single axis is divided into smaller equal sub-parts which are then reconstructed using edge preserved kriging interpolation to predict the missing slice information. In order to speed up the process of interpolation, we have used multiprocessing by carrying out the initial interpolation on parallel cores. From the 3D matrix thus formed, shearlet transform is applied to estimate the edges considering the 2D blocks along the $Z$ axis, and to minimize the blurring effect using a proposed mean-median logic. Finally, for visualization, the sub-matrices are merged into a final 3D matrix. Next, the newly formed 3D matrix is split up into voxels and marching cubes method is applied to get the approximate 3D image for viewing. To the best of our knowledge it is a first of its kind approach based on kriging interpolation and multiprocessing for 3D reconstruction from 2D slices, and approximately 98.89\% accuracy is achieved with respect to similarity metrics for image comparison. The time required for reconstruction has also been reduced by approximately 70\% with multiprocessing even for a large input data set compared to that with single core processing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge