Soheil Soltani
Prostate cancer histopathology with label-free multispectral deep UV microscopy quantifies phenotypes of tumor grade and aggressiveness
Jun 01, 2021


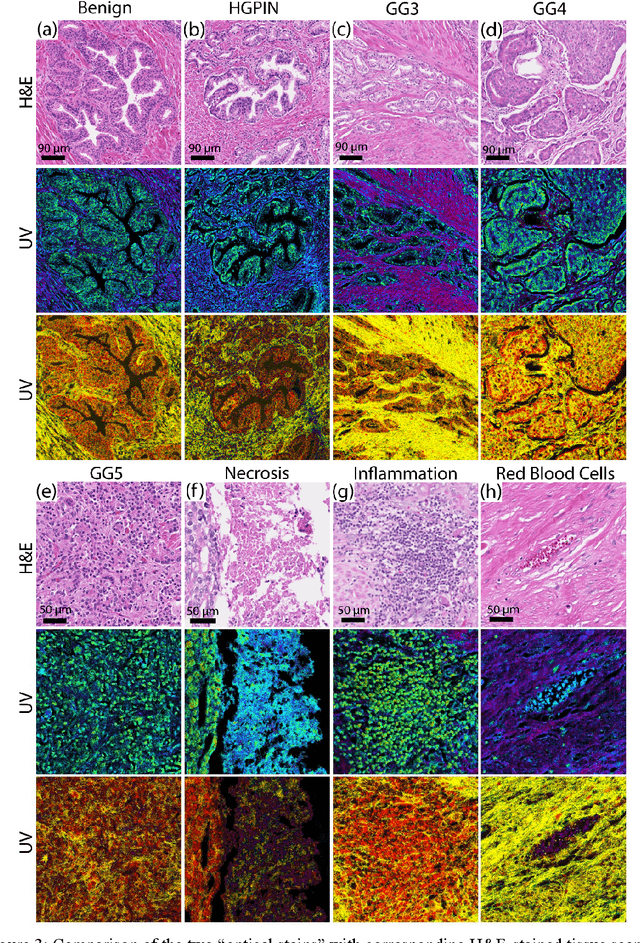
Abstract:Identifying prostate cancer patients that are harboring aggressive forms of prostate cancer remains a significant clinical challenge. To shed light on this problem, we develop an approach based on multispectral deep-ultraviolet (UV) microscopy that provides novel quantitative insight into the aggressiveness and grade of this disease. First, we find that UV spectral signatures from endogenous molecules give rise to a phenotypical continuum that differentiates critical structures of thin tissue sections with subcellular spatial resolution, including nuclei, cytoplasm, stroma, basal cells, nerves, and inflammation. Further, we show that this phenotypical continuum can be applied as a surrogate biomarker of prostate cancer malignancy, where patients with the most aggressive tumors show a ubiquitous glandular phenotypical shift. Lastly, we adapt a two-part Cycle-consistent Generative Adversarial Network to translate the label-free deep-UV images into virtual hematoxylin and eosin (H&E) stained images. Agreement between the virtual H&E images and the gold standard H&E-stained tissue sections is evaluated by a panel of pathologists who find that the two modalities are in excellent agreement. This work has significant implications towards improving our ability to objectively quantify prostate cancer grade and aggressiveness, thus improving the management and clinical outcomes of prostate cancer patients. This same approach can also be applied broadly in other tumor types to achieve low-cost, stain-free, quantitative histopathological analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge