Simon Chun-Ho Yu
Unsupervised domain adaptation for cross-modality liver segmentation via joint adversarial learning and self-learning
Sep 20, 2021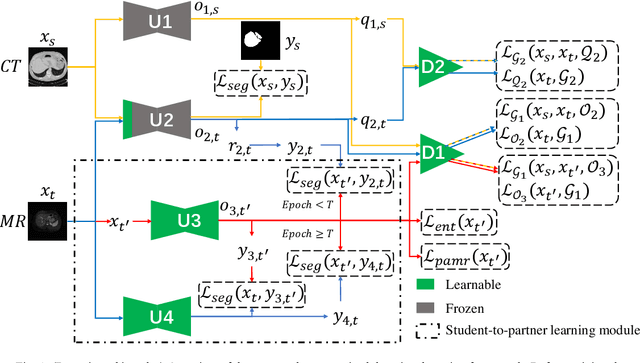
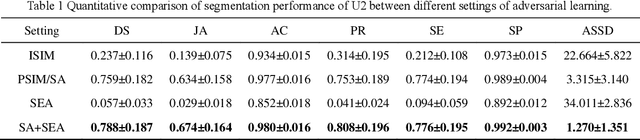
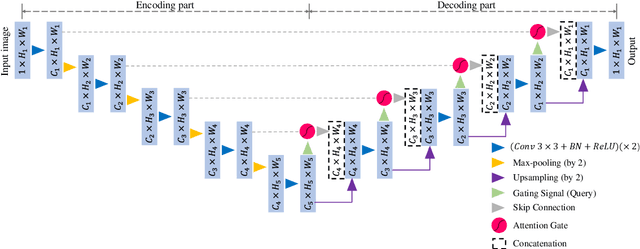
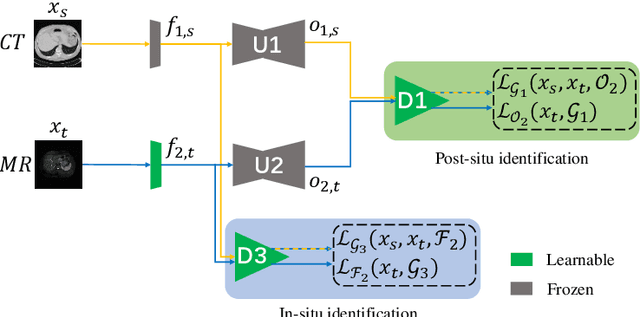
Abstract:Liver segmentation on images acquired using computed tomography (CT) and magnetic resonance imaging (MRI) plays an important role in clinical management of liver diseases. Compared to MRI, CT images of liver are more abundant and readily available. However, MRI can provide richer quantitative information of the liver compared to CT. Thus, it is desirable to achieve unsupervised domain adaptation for transferring the learned knowledge from the source domain containing labeled CT images to the target domain containing unlabeled MR images. In this work, we report a novel unsupervised domain adaptation framework for cross-modality liver segmentation via joint adversarial learning and self-learning. We propose joint semantic-aware and shape-entropy-aware adversarial learning with post-situ identification manner to implicitly align the distribution of task-related features extracted from the target domain with those from the source domain. In proposed framework, a network is trained with the above two adversarial losses in an unsupervised manner, and then a mean completer of pseudo-label generation is employed to produce pseudo-labels to train the next network (desired model). Additionally, semantic-aware adversarial learning and two self-learning methods, including pixel-adaptive mask refinement and student-to-partner learning, are proposed to train the desired model. To improve the robustness of the desired model, a low-signal augmentation function is proposed to transform MRI images as the input of the desired model to handle hard samples. Using the public data sets, our experiments demonstrated the proposed unsupervised domain adaptation framework outperformed four supervised learning methods with a Dice score 0.912 plus or minus 0.037 (mean plus or minus standard deviation).
Context-aware PolyUNet for Liver and Lesion Segmentation from Abdominal CT Images
Jun 21, 2021



Abstract:Accurate liver and lesion segmentation from computed tomography (CT) images are highly demanded in clinical practice for assisting the diagnosis and assessment of hepatic tumor disease. However, automatic liver and lesion segmentation from contrast-enhanced CT volumes is extremely challenging due to the diversity in contrast, resolution, and quality of images. Previous methods based on UNet for 2D slice-by-slice or 3D volume-by-volume segmentation either lack sufficient spatial contexts or suffer from high GPU computational cost, which limits the performance. To tackle these issues, we propose a novel context-aware PolyUNet for accurate liver and lesion segmentation. It jointly explores structural diversity and consecutive t-adjacent slices to enrich feature expressive power and spatial contextual information while avoiding the overload of GPU memory consumption. In addition, we utilize zoom out/in and two-stage refinement strategy to exclude the irrelevant contexts and focus on the specific region for the fine-grained segmentation. Our method achieved very competitive performance at the MICCAI 2017 Liver Tumor Segmentation (LiTS) Challenge among all tasks with a single model and ranked the $3^{rd}$, $12^{th}$, $2^{nd}$, and $5^{th}$ places in the liver segmentation, lesion segmentation, lesion detection, and tumor burden estimation, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge