Siamac Fazli
A Review of Modern Approaches for Coronary Angiography Imaging Analysis
Sep 28, 2022



Abstract:Coronary Heart Disease (CHD) is a leading cause of death in the modern world. The development of modern analytical tools for diagnostics and treatment of CHD is receiving substantial attention from the scientific community. Deep learning-based algorithms, such as segmentation networks and detectors, play an important role in assisting medical professionals by providing timely analysis of a patient's angiograms. This paper focuses on X-Ray Coronary Angiography (XCA), which is considered to be a "gold standard" in the diagnosis and treatment of CHD. First, we describe publicly available datasets of XCA images. Then, classical and modern techniques of image preprocessing are reviewed. In addition, common frame selection techniques are discussed, which are an important factor of input quality and thus model performance. In the following two chapters we discuss modern vessel segmentation and stenosis detection networks and, finally, open problems and current limitations of the current state-of-the-art.
MLT-LE: predicting drug-target binding affinity with multi-task residual neural networks
Sep 13, 2022

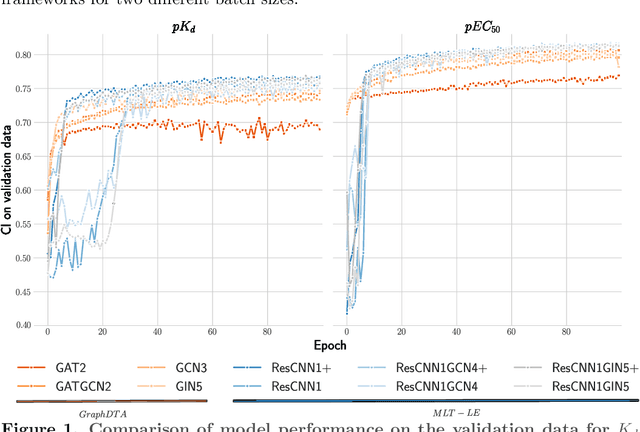
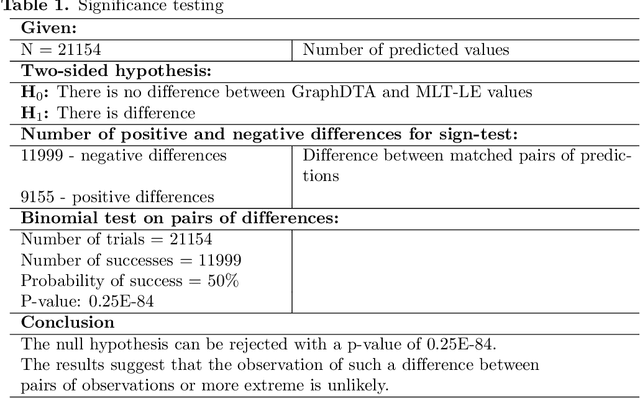
Abstract:Assessing drug-target affinity is a critical step in the drug discovery and development process, but to obtain such data experimentally is both time consuming and expensive. For this reason, computational methods for predicting binding strength are being widely developed. However, these methods typically use a single-task approach for prediction, thus ignoring the additional information that can be extracted from the data and used to drive the learning process. Thereafter in this work, we present a multi-task approach for binding strength prediction. Our results suggest that these prediction can indeed benefit from a multi-task learning approach, by utilizing added information from related tasks and multi-task induced regularization.
A biologically-inspired evaluation of molecular generative machine learning
Aug 20, 2022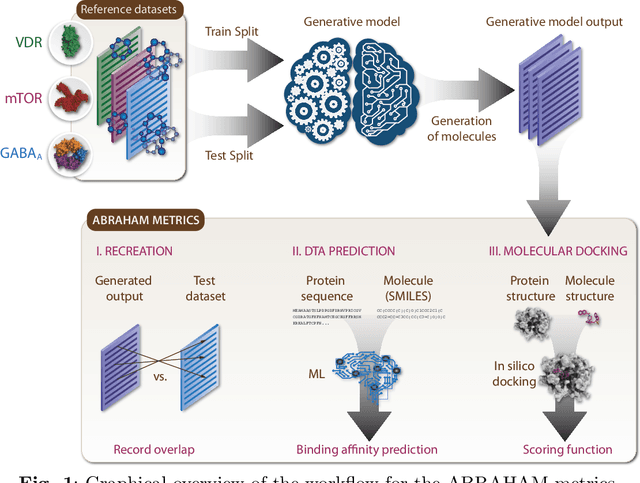
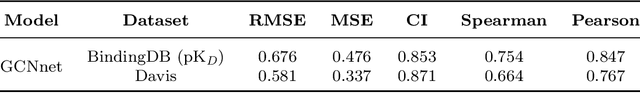
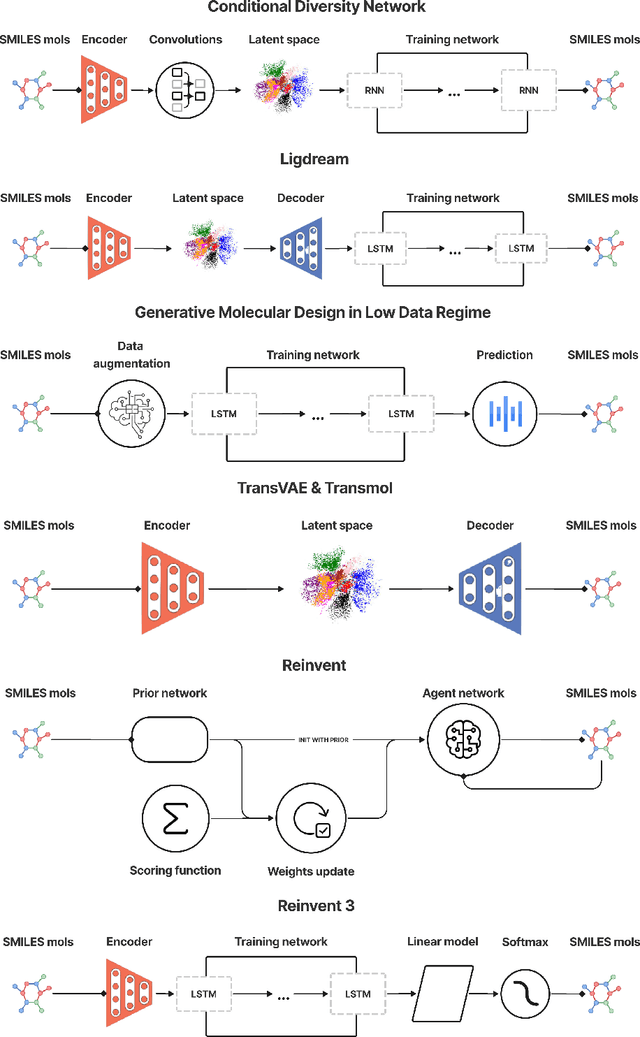
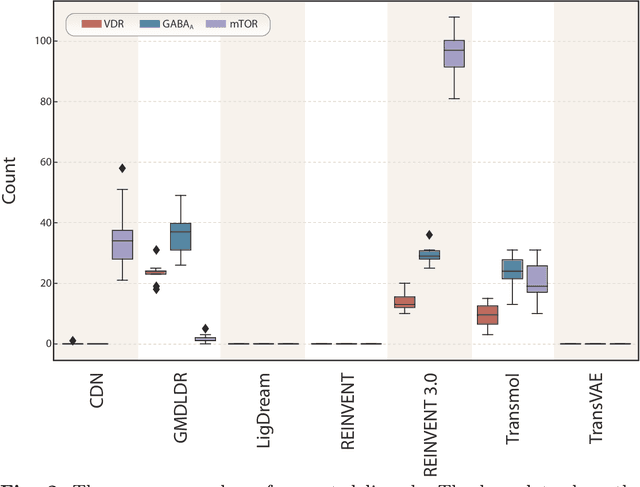
Abstract:While generative models have recently become ubiquitous in many scientific areas, less attention has been paid to their evaluation. For molecular generative models, the state-of-the-art examines their output in isolation or in relation to its input. However, their biological and functional properties, such as ligand-target interaction is not being addressed. In this study, a novel biologically-inspired benchmark for the evaluation of molecular generative models is proposed. Specifically, three diverse reference datasets are designed and a set of metrics are introduced which are directly relevant to the drug discovery process. In particular we propose a recreation metric, apply drug-target affinity prediction and molecular docking as complementary techniques for the evaluation of generative outputs. While all three metrics show consistent results across the tested generative models, a more detailed comparison of drug-target affinity binding and molecular docking scores revealed that unimodal predictiors can lead to erroneous conclusions about target binding on a molecular level and a multi-modal approach is thus preferrable. The key advantage of this framework is that it incorporates prior physico-chemical domain knowledge into the benchmarking process by focusing explicitly on ligand-target interactions and thus creating a highly efficient tool not only for evaluating molecular generative outputs in particular, but also for enriching the drug discovery process in general.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge