Shameek Ghosh
Understanding patient complaint characteristics using contextual clinical BERT embeddings
Feb 14, 2020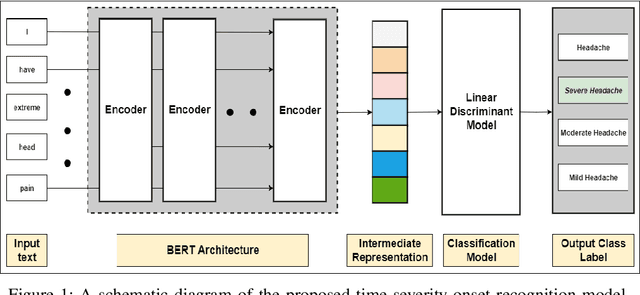
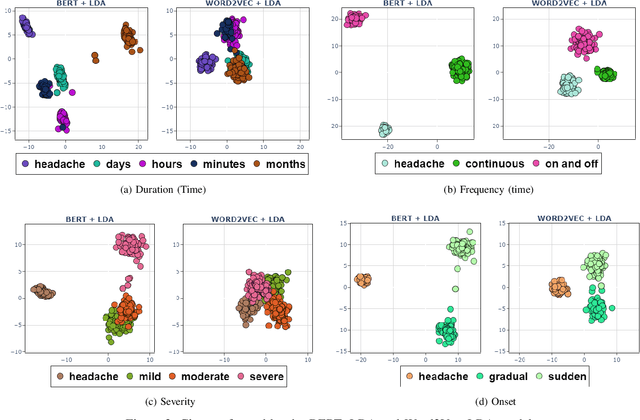
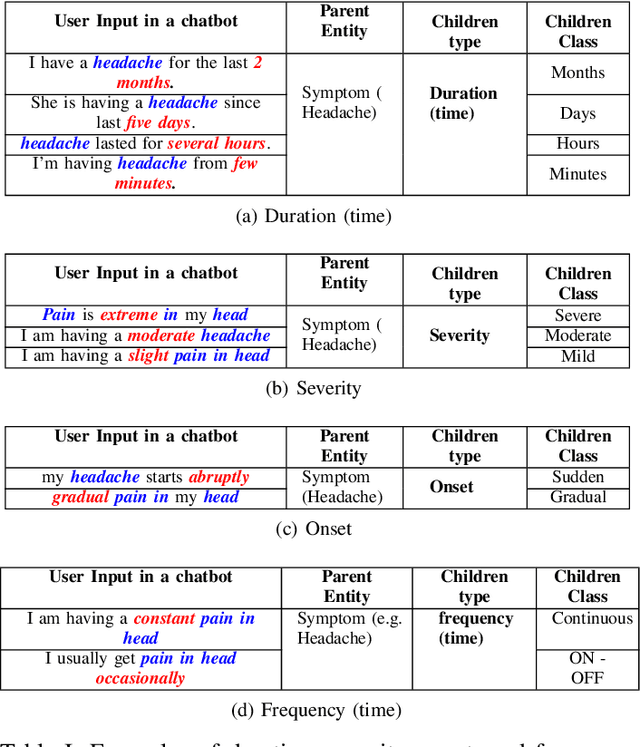
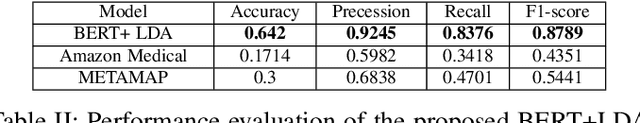
Abstract:In clinical conversational applications, extracted entities tend to capture the main subject of a patient's complaint, namely symptoms or diseases. However, they mostly fail to recognize the characterizations of a complaint such as the time, the onset, and the severity. For example, if the input is "I have a headache and it is extreme", state-of-the-art models only recognize the main symptom entity - headache, but ignore the severity factor of "extreme", that characterizes headache. In this paper, we design a two-stage approach to detect the characterizations of entities like symptoms presented by general users in contexts where they would describe their symptoms to a clinician. We use Word2Vec and BERT to encode clinical text given by the patients. We transform the output and re-frame the task as multi-label classification problem. Finally, we combine the processed encodings with the Linear Discriminant Analysis (LDA) algorithm to classify the characterizations of the main entity. Experimental results demonstrate that our method achieves 40-50% improvement on the accuracy over the state-of-the-art models.
Biogeography-Based Informative Gene Selection and Cancer Classification Using SVM and Random Forests
Jul 12, 2012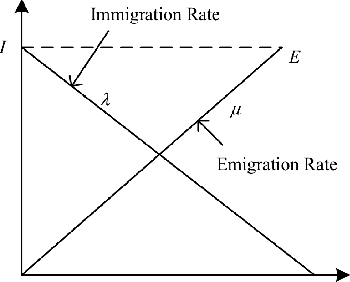
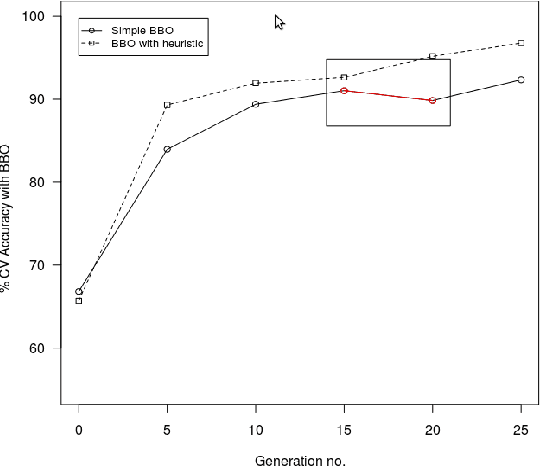
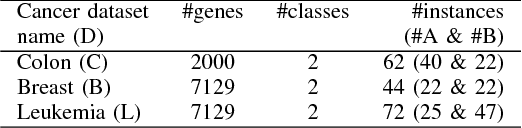
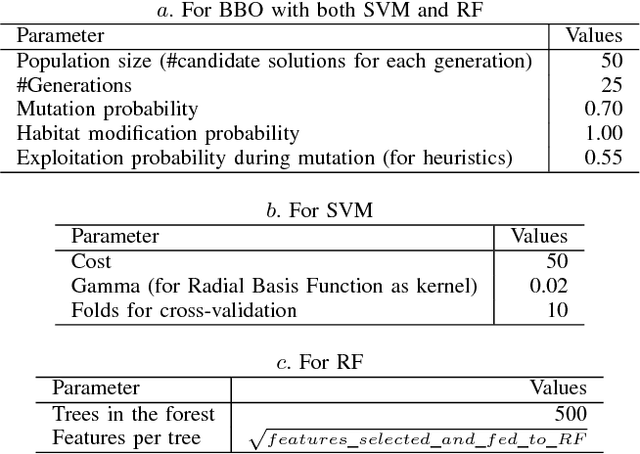
Abstract:Microarray cancer gene expression data comprise of very high dimensions. Reducing the dimensions helps in improving the overall analysis and classification performance. We propose two hybrid techniques, Biogeography - based Optimization - Random Forests (BBO - RF) and BBO - SVM (Support Vector Machines) with gene ranking as a heuristic, for microarray gene expression analysis. This heuristic is obtained from information gain filter ranking procedure. The BBO algorithm generates a population of candidate subset of genes, as part of an ecosystem of habitats, and employs the migration and mutation processes across multiple generations of the population to improve the classification accuracy. The fitness of each gene subset is assessed by the classifiers - SVM and Random Forests. The performances of these hybrid techniques are evaluated on three cancer gene expression datasets retrieved from the Kent Ridge Biomedical datasets collection and the libSVM data repository. Our results demonstrate that genes selected by the proposed techniques yield classification accuracies comparable to previously reported algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge