Shahryar Khorasani
Two-sample Testing Using Deep Learning
Oct 14, 2019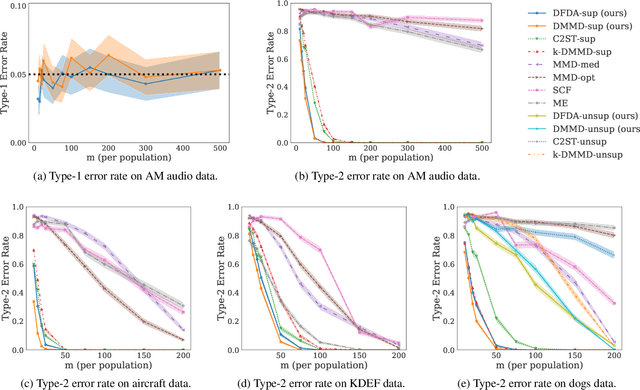
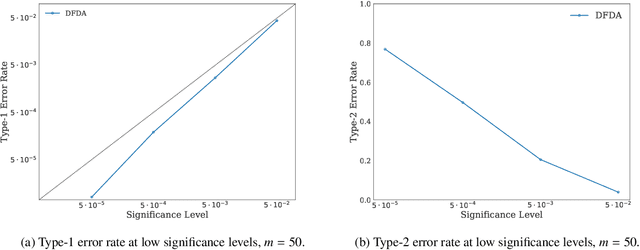
Abstract:We propose a two-sample testing procedure based on learned deep neural network representations. To this end, we define two test statistics that perform an asymptotic location test on data samples mapped onto a hidden layer. The tests are consistent and asymptotically control the type-1 error rate. Their test statistics can be evaluated in linear time (in the sample size). Suitable data representations are obtained in a data-driven way, by solving a supervised or unsupervised transfer-learning task on an auxiliary (potentially distinct) data set. If no auxiliary data is available, we split the data into two chunks: one for learning representations and one for computing the test statistic. In experiments on audio samples, natural images and three-dimensional neuroimaging data our tests yield significant decreases in type-2 error rate (up to 35 percentage points) compared to state-of-the-art two-sample tests such as kernel-methods and classifier two-sample tests.
Integrating omics and MRI data with kernel-based tests and CNNs to identify rare genetic markers for Alzheimer's disease
Dec 02, 2018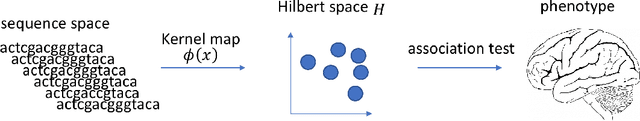
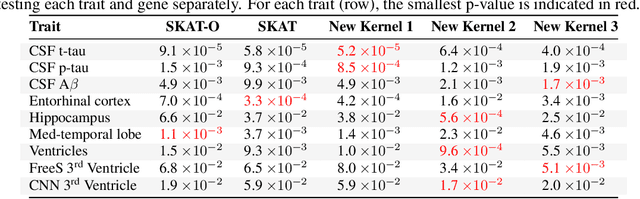
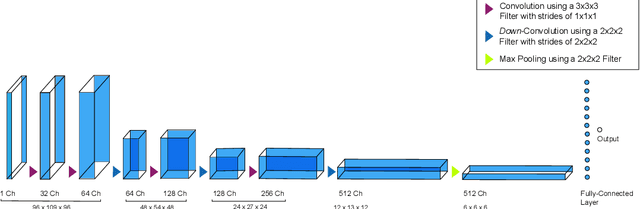
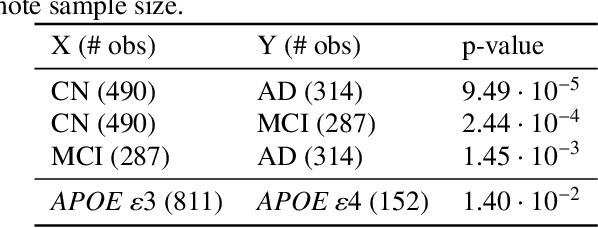
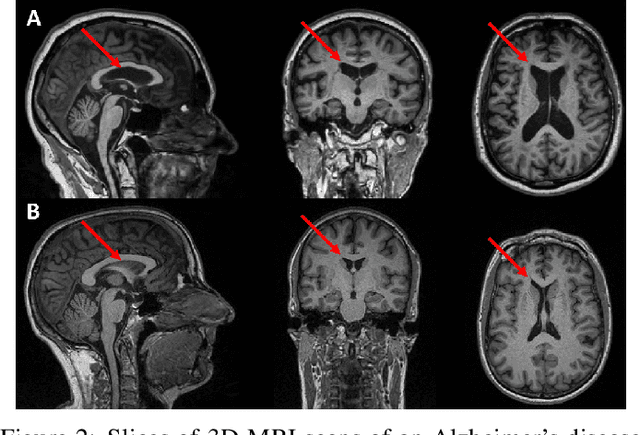
Abstract:For precision medicine and personalized treatment, we need to identify predictive markers of disease. We focus on Alzheimer's disease (AD), where magnetic resonance imaging scans provide information about the disease status. By combining imaging with genome sequencing, we aim at identifying rare genetic markers associated with quantitative traits predicted from convolutional neural networks (CNNs), which traditionally have been derived manually by experts. Kernel-based tests are a powerful tool for associating sets of genetic variants, but how to optimally model rare genetic variants is still an open research question. We propose a generalized set of kernels that incorporate prior information from various annotations and multi-omics data. In the analysis of data from the Alzheimer's Disease Neuroimaging Initiative (ADNI), we evaluate whether (i) CNNs yield precise and reliable brain traits, and (ii) the novel kernel-based tests can help to identify loci associated with AD. The results indicate that CNNs provide a fast, scalable and precise tool to derive quantitative AD traits and that new kernels integrating domain knowledge can yield higher power in association tests of very rare variants.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge