Seth A. Faith
Global forensic geolocation with deep neural networks
May 28, 2019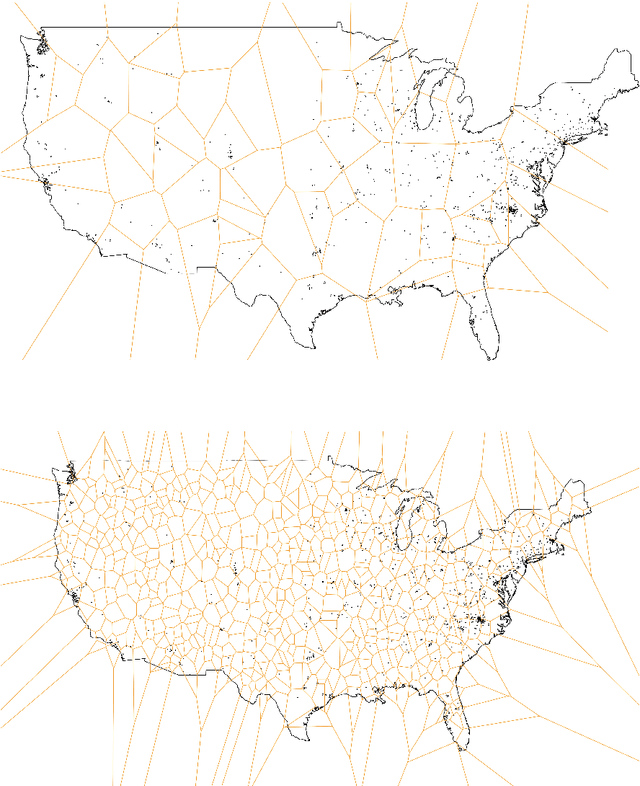
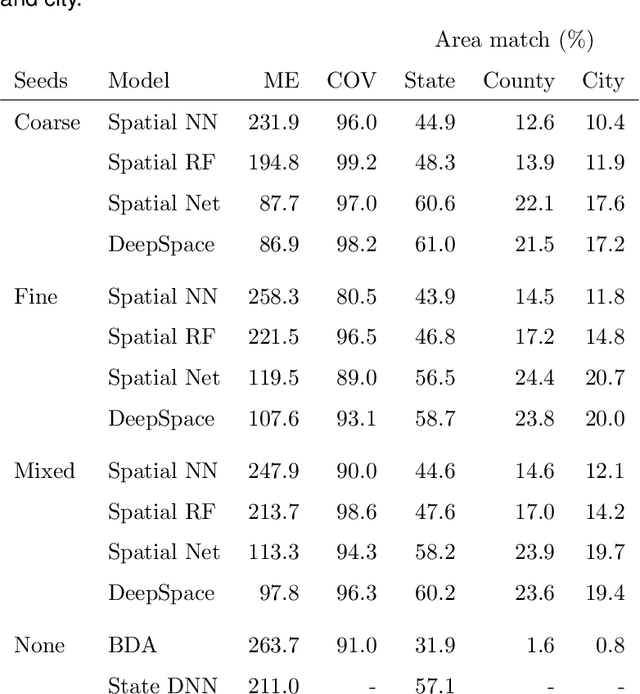
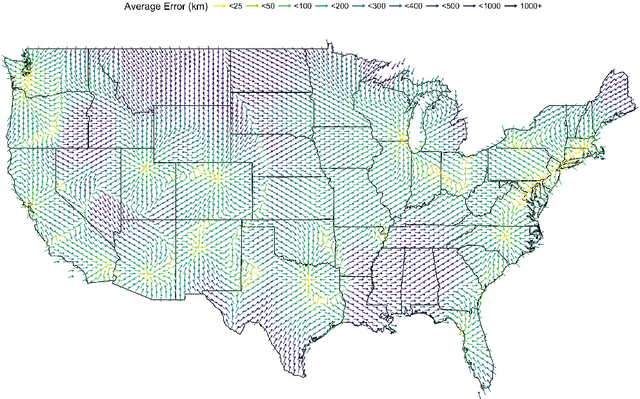
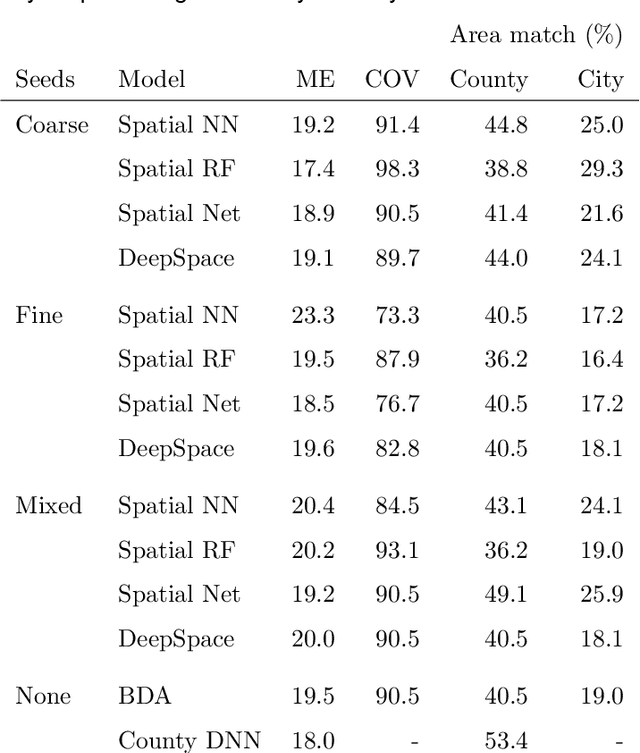
Abstract:An important problem in forensic analyses is identifying the provenance of materials at a crime scene, such as biological material on a piece of clothing. This procedure, known as geolocation, is conventionally guided by expert knowledge of the biological evidence and therefore tends to be application-specific, labor-intensive, and subjective. Purely data-driven methods have yet to be fully realized due in part to the lack of a sufficiently rich data source. However, high-throughput sequencing technologies are able to identify tens of thousands of microbial taxa using DNA recovered from a single swab collected from nearly any object or surface. We present a new algorithm for geolocation that aggregates over an ensemble of deep neural network classifiers trained on randomly-generated Voronoi partitions of a spatial domain. We apply the algorithm to fungi present in each of 1300 dust samples collected across the continental United States and then to a global dataset of dust samples from 28 countries. Our algorithm makes remarkably good point predictions with more than half of the geolocation errors under 100 kilometers for the continental analysis and nearly 90% classification accuracy of a sample's country of origin for the global analysis. We suggest that the effectiveness of this model sets the stage for a new, quantitative approach to forensic geolocation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge