Sehwan Moon
School of Electrical Engineering and Computer Science of Gwangju Institute of Science and Technology
DepressLLM: Interpretable domain-adapted language model for depression detection from real-world narratives
Aug 12, 2025Abstract:Advances in large language models (LLMs) have enabled a wide range of applications. However, depression prediction is hindered by the lack of large-scale, high-quality, and rigorously annotated datasets. This study introduces DepressLLM, trained and evaluated on a novel corpus of 3,699 autobiographical narratives reflecting both happiness and distress. DepressLLM provides interpretable depression predictions and, via its Score-guided Token Probability Summation (SToPS) module, delivers both improved classification performance and reliable confidence estimates, achieving an AUC of 0.789, which rises to 0.904 on samples with confidence $\geq$ 0.95. To validate its robustness to heterogeneous data, we evaluated DepressLLM on in-house datasets, including an Ecological Momentary Assessment (EMA) corpus of daily stress and mood recordings, and on public clinical interview data. Finally, a psychiatric review of high-confidence misclassifications highlighted key model and data limitations that suggest directions for future refinements. These findings demonstrate that interpretable AI can enable earlier diagnosis of depression and underscore the promise of medical AI in psychiatry.
Deep Metric Loss for Multimodal Learning
Aug 21, 2023Abstract:Multimodal learning often outperforms its unimodal counterparts by exploiting unimodal contributions and cross-modal interactions. However, focusing only on integrating multimodal features into a unified comprehensive representation overlooks the unimodal characteristics. In real data, the contributions of modalities can vary from instance to instance, and they often reinforce or conflict with each other. In this study, we introduce a novel \text{MultiModal} loss paradigm for multimodal learning, which subgroups instances according to their unimodal contributions. \text{MultiModal} loss can prevent inefficient learning caused by overfitting and efficiently optimize multimodal models. On synthetic data, \text{MultiModal} loss demonstrates improved classification performance by subgrouping difficult instances within certain modalities. On four real multimodal datasets, our loss is empirically shown to improve the performance of recent models. Ablation studies verify the effectiveness of our loss. Additionally, we show that our loss generates a reliable prediction score for each modality, which is essential for subgrouping. Our \text{MultiModal} loss is a novel loss function to subgroup instances according to the contribution of modalities in multimodal learning and is applicable to a variety of multimodal models with unimodal decisions. Our code is available at https://github.com/SehwanMoon/MultiModalLoss.
SDGCCA: Supervised Deep Generalized Canonical Correlation Analysis for Multi-omics Integration
Apr 17, 2022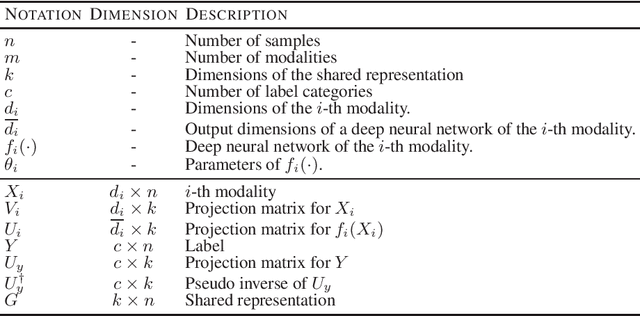
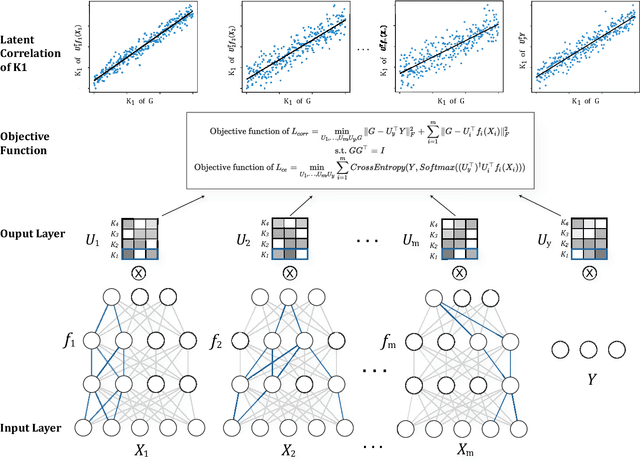
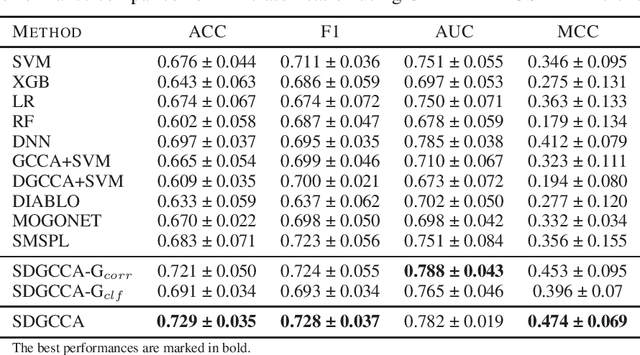
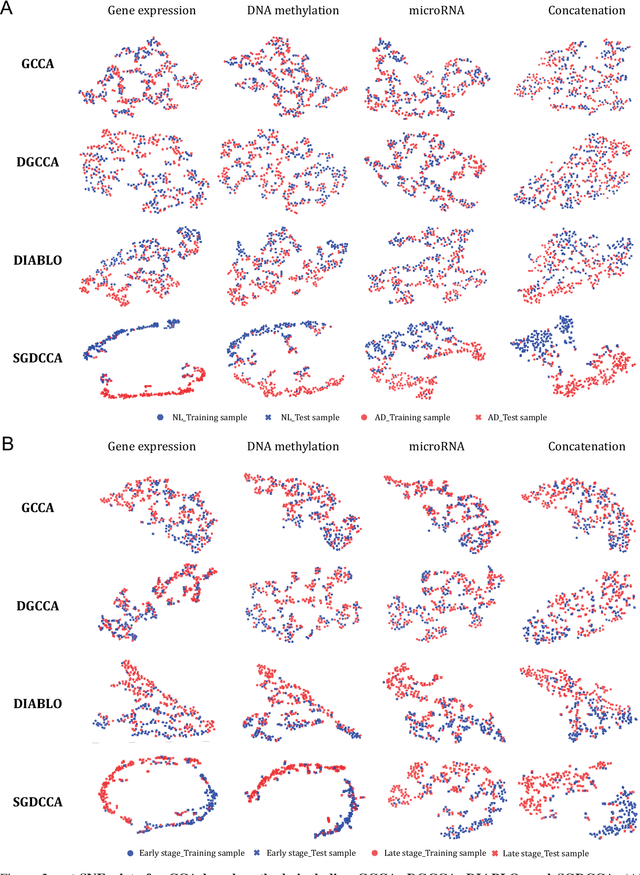
Abstract:Integration of multi-omics data provides opportunities for revealing biological mechanisms related to certain phenotypes. We propose a novel method of multi-omics integration called supervised deep generalized canonical correlation analysis (SDGCCA) for modeling correlation structures between nonlinear multi-omics manifolds, aiming for improving classification of phenotypes and revealing biomarkers related to phenotypes. SDGCCA addresses the limitations of other canonical correlation analysis (CCA)-based models (e.g., deep CCA, deep generalized CCA) by considering complex/nonlinear cross-data correlations and discriminating phenotype groups. Although there are a few methods for nonlinear CCA projections for discriminant purposes of phenotypes, they only consider two views. On the other hand, SDGCCA is the nonlinear multiview CCA projection method for discrimination. When we applied SDGCCA to prediction of patients of Alzheimer's disease (AD) and discrimination of early- and late-stage cancers, it outperformed other CCA-based methods and other supervised methods. In addition, we demonstrate that SDGCCA can be used for feature selection to identify important multi-omics biomarkers. In the application on AD data, SDGCCA identified clusters of genes in multi-omics data, which are well known to be associated with AD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge