Sandrine Prost
Enabling Progressive Whole-slide Image Analysis with Multi-scale Pyramidal Network
Feb 02, 2026Abstract:Multiple-instance Learning (MIL) is commonly used to undertake computational pathology (CPath) tasks, and the use of multi-scale patches allows diverse features across scales to be learned. Previous studies using multi-scale features in clinical applications rely on multiple inputs across magnifications with late feature fusion, which does not retain the link between features across scales while the inputs are dependent on arbitrary, manufacturer-defined magnifications, being inflexible and computationally expensive. In this paper, we propose the Multi-scale Pyramidal Network (MSPN), which is plug-and-play over attention-based MIL that introduces progressive multi-scale analysis on WSI. Our MSPN consists of (1) grid-based remapping that uses high magnification features to derive coarse features and (2) the coarse guidance network (CGN) that learns coarse contexts. We benchmark MSPN as an add-on module to 4 attention-based frameworks using 4 clinically relevant tasks across 3 types of foundation model, as well as the pre-trained MIL framework. We show that MSPN consistently improves MIL across the compared configurations and tasks, while being lightweight and easy-to-use.
Multiple Instance Learning with Coarse-to-Fine Self-Distillation
Feb 04, 2025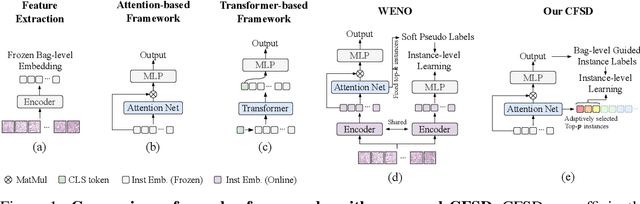
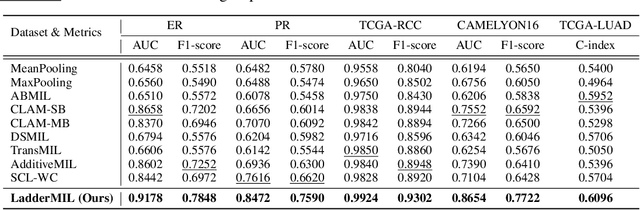
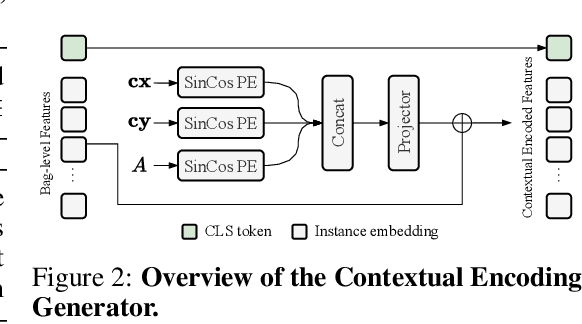
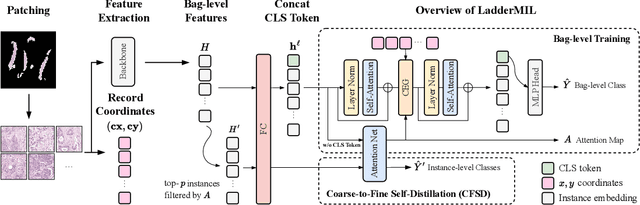
Abstract:Multiple Instance Learning (MIL) for whole slide image (WSI) analysis in computational pathology often neglects instance-level learning as supervision is typically provided only at the bag level. In this work, we present PathMIL, a framework designed to improve MIL through two perspectives: (1) employing instance-level supervision and (2) learning inter-instance contextual information on bag level. Firstly, we propose a novel Coarse-to-Fine Self-Distillation (CFSD) paradigm, to probe and distil a classifier trained with bag-level information to obtain instance-level labels which could effectively provide the supervision for the same classifier in a finer way. Secondly, to capture inter-instance contextual information in WSI, we propose Two-Dimensional Positional Encoding (2DPE), which encodes the spatial appearance of instances within a bag. We also theoretically and empirically prove the instance-level learnability of CFSD. PathMIL is evaluated on multiple benchmarking tasks, including subtype classification (TCGA-NSCLC), tumour classification (CAMELYON16), and an internal benchmark for breast cancer receptor status classification. Our method achieves state-of-the-art performance, with AUC scores of 0.9152 and 0.8524 for estrogen and progesterone receptor status classification, respectively, an AUC of 0.9618 for subtype classification, and 0.8634 for tumour classification, surpassing existing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge