S. Klein
Multi-Atlas Segmentation and Spatial Alignment of the Human Embryo in First Trimester 3D Ultrasound
Feb 14, 2022



Abstract:Segmentation and spatial alignment of ultrasound (US) imaging data acquired in the in first trimester are crucial for monitoring human embryonic growth and development throughout this crucial period of life. Current approaches are either manual or semi-automatic and are therefore very time-consuming and prone to errors. To automate these tasks, we propose a multi-atlas framework for automatic segmentation and spatial alignment of the embryo using deep learning with minimal supervision. Our framework learns to register the embryo to an atlas, which consists of the US images acquired at a range of gestational age (GA), segmented and spatially aligned to a predefined standard orientation. From this, we can derive the segmentation of the embryo and put the embryo in standard orientation. US images acquired at 8+0 till 12+6 weeks GA were used and eight pregnancies were selected as atlas images. We evaluated different fusion strategies to incorporate multiple atlases: 1) training the framework using atlas images from a single pregnancy, 2) training the framework with data of all available atlases and 3) ensembling of the frameworks trained per pregnancy. To evaluate the performance, we calculated the Dice score over the test set. We found that training the framework using all available atlases outperformed ensembling and gave similar results compared to the best of all frameworks trained on a single subject. Furthermore, we found that selecting images from the four atlases closest in GA out of all available atlases, regardless of the individual quality, gave the best results with a median Dice score of 0.72. We conclude that our framework can accurately segment and spatially align the embryo in first trimester 3D US images and is robust for the variation in quality that existed in the available atlases. Our code is publicly available at: https://github.com/wapbastiaansen/multi-atlas-seg-reg.
Recurrent Inference Machines as inverse problem solvers for MR relaxometry
Jun 08, 2021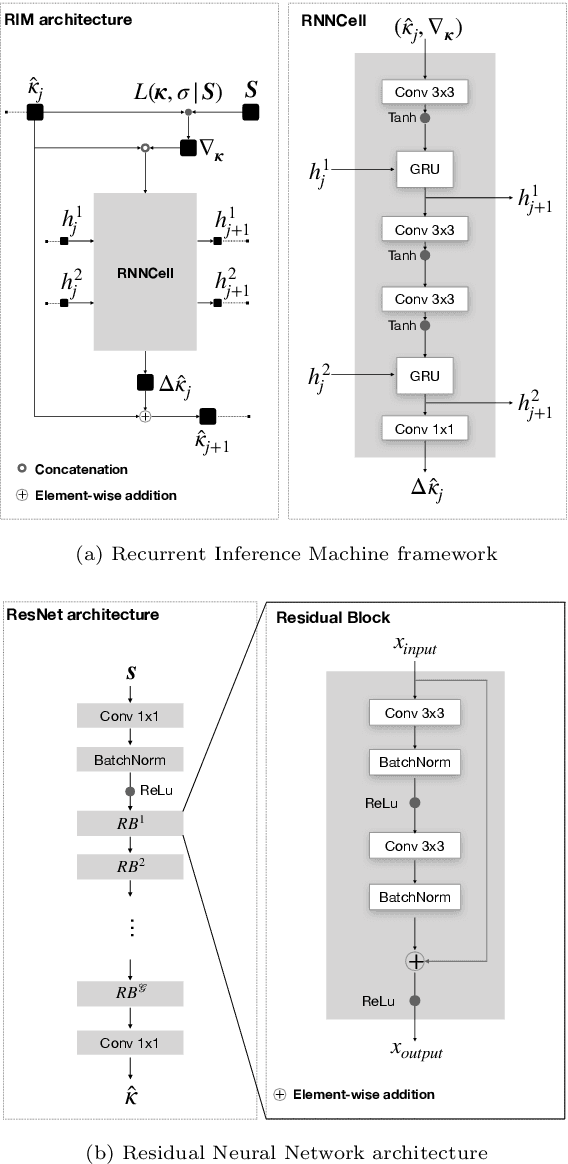
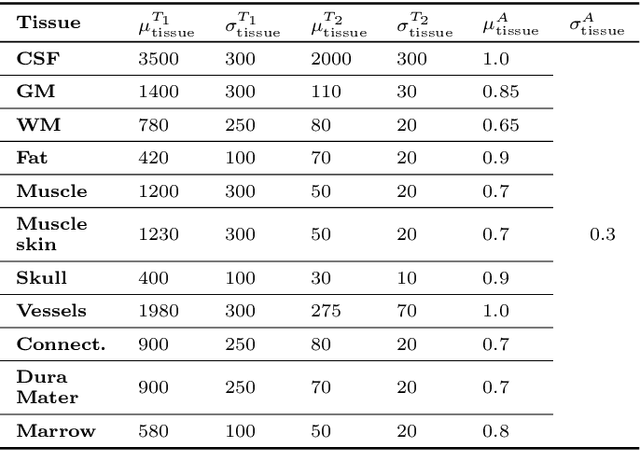
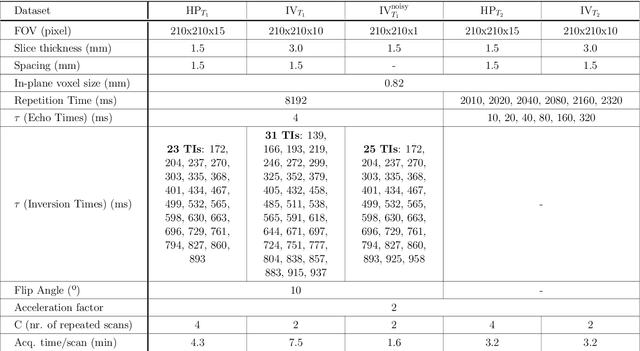
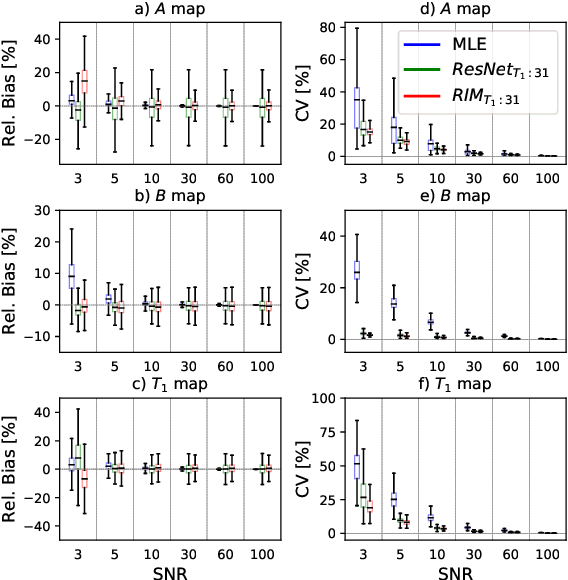
Abstract:In this paper, we propose the use of Recurrent Inference Machines (RIMs) to perform T1 and T2 mapping. The RIM is a neural network framework that learns an iterative inference process based on the signal model, similar to conventional statistical methods for quantitative MRI (QMRI), such as the Maximum Likelihood Estimator (MLE). This framework combines the advantages of both data-driven and model-based methods, and, we hypothesize, is a promising tool for QMRI. Previously, RIMs were used to solve linear inverse reconstruction problems. Here, we show that they can also be used to optimize non-linear problems and estimate relaxometry maps with high precision and accuracy. The developed RIM framework is evaluated in terms of accuracy and precision and compared to an MLE method and an implementation of the ResNet. The results show that the RIM improves the quality of estimates compared to the other techniques in Monte Carlo experiments with simulated data, test-retest analysis of a system phantom, and in-vivo scans. Additionally, inference with the RIM is 150 times faster than the MLE, and robustness to (slight) variations of scanning parameters is demonstrated. Hence, the RIM is a promising and flexible method for QMRI. Coupled with an open-source training data generation tool, it presents a compelling alternative to previous methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge