Ruben Kruiper
SPaR.txt, a cheap Shallow Parsing approach for Regulatory texts
Oct 04, 2021
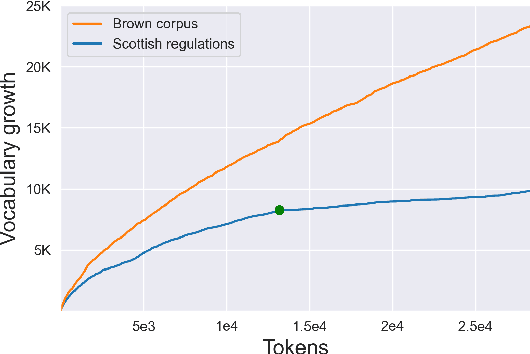
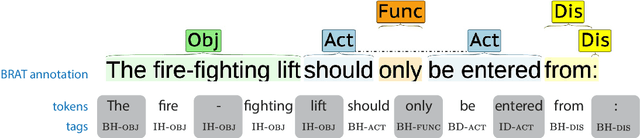
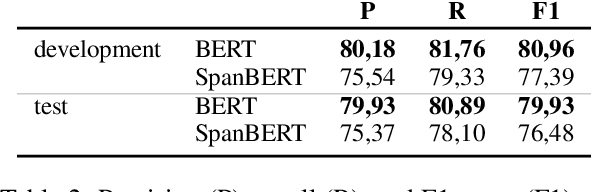
Abstract:Automated Compliance Checking (ACC) systems aim to semantically parse building regulations to a set of rules. However, semantic parsing is known to be hard and requires large amounts of training data. The complexity of creating such training data has led to research that focuses on small sub-tasks, such as shallow parsing or the extraction of a limited subset of rules. This study introduces a shallow parsing task for which training data is relatively cheap to create, with the aim of learning a lexicon for ACC. We annotate a small domain-specific dataset of 200 sentences, SPaR.txt, and train a sequence tagger that achieves 79,93 F1-score on the test set. We then show through manual evaluation that the model identifies most (89,84%) defined terms in a set of building regulation documents, and that both contiguous and discontiguous Multi-Word Expressions (MWE) are discovered with reasonable accuracy (70,3%).
In Layman's Terms: Semi-Open Relation Extraction from Scientific Texts
May 26, 2020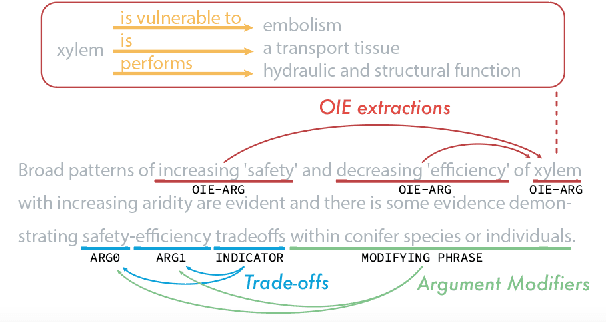
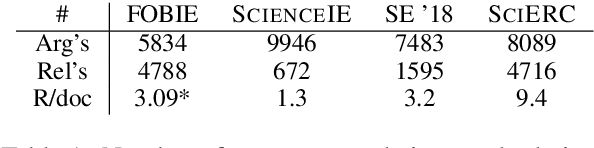
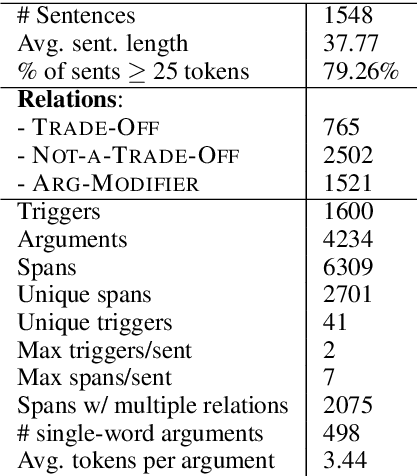
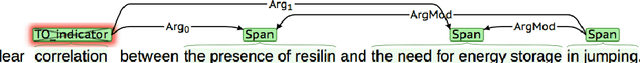
Abstract:Information Extraction (IE) from scientific texts can be used to guide readers to the central information in scientific documents. But narrow IE systems extract only a fraction of the information captured, and Open IE systems do not perform well on the long and complex sentences encountered in scientific texts. In this work we combine the output of both types of systems to achieve Semi-Open Relation Extraction, a new task that we explore in the Biology domain. First, we present the Focused Open Biological Information Extraction (FOBIE) dataset and use FOBIE to train a state-of-the-art narrow scientific IE system to extract trade-off relations and arguments that are central to biology texts. We then run both the narrow IE system and a state-of-the-art Open IE system on a corpus of 10k open-access scientific biological texts. We show that a significant amount (65%) of erroneous and uninformative Open IE extractions can be filtered using narrow IE extractions. Furthermore, we show that the retained extractions are significantly more often informative to a reader.
A Scientific Information Extraction Dataset for Nature Inspired Engineering
May 26, 2020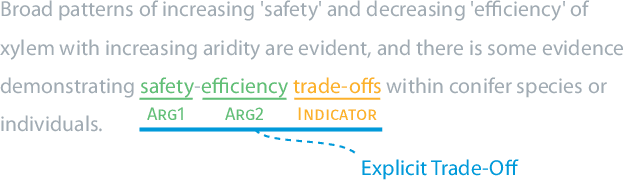
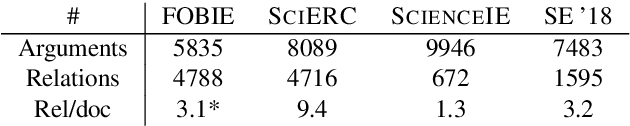

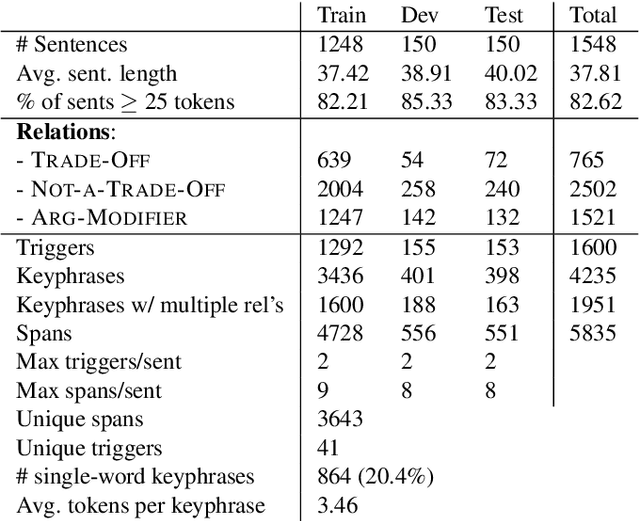
Abstract:Nature has inspired various ground-breaking technological developments in applications ranging from robotics to aerospace engineering and the manufacturing of medical devices. However, accessing the information captured in scientific biology texts is a time-consuming and hard task that requires domain-specific knowledge. Improving access for outsiders can help interdisciplinary research like Nature Inspired Engineering. This paper describes a dataset of 1,500 manually-annotated sentences that express domain-independent relations between central concepts in a scientific biology text, such as trade-offs and correlations. The arguments of these relations can be Multi Word Expressions and have been annotated with modifying phrases to form non-projective graphs. The dataset allows for training and evaluating Relation Extraction algorithms that aim for coarse-grained typing of scientific biological documents, enabling a high-level filter for engineers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge