Rosemary He
Statistical testing on generative AI anomaly detection tools in Alzheimer's Disease diagnosis
Oct 17, 2024

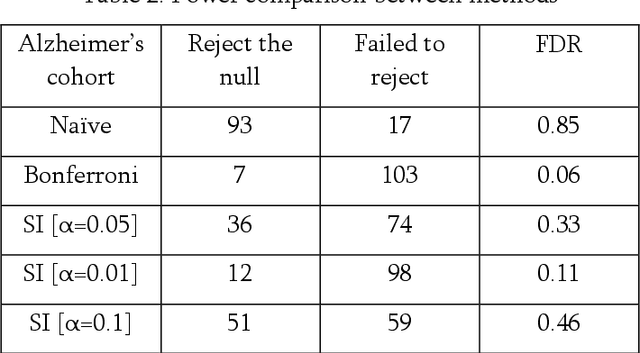
Abstract:Alzheimer's Disease is challenging to diagnose due to our limited understanding of its mechanism and large heterogeneity among patients. Neurodegeneration is studied widely as a biomarker for clinical diagnosis, which can be measured from time series MRI progression. On the other hand, generative AI has shown promise in anomaly detection in medical imaging and used for tasks including tumor detection. However, testing the reliability of such data-driven methods is non-trivial due to the issue of double-dipping in hypothesis testing. In this work, we propose to solve this issue with selective inference and develop a reliable generative AI method for Alzheimer's prediction. We show that compared to traditional statistical methods with highly inflated p-values, selective inference successfully controls the false discovery rate under the desired alpha level while retaining statistical power. In practice, our pipeline could assist clinicians in Alzheimer's diagnosis and early intervention.
Individualized multi-horizon MRI trajectory prediction for Alzheimer's Disease
Aug 04, 2024



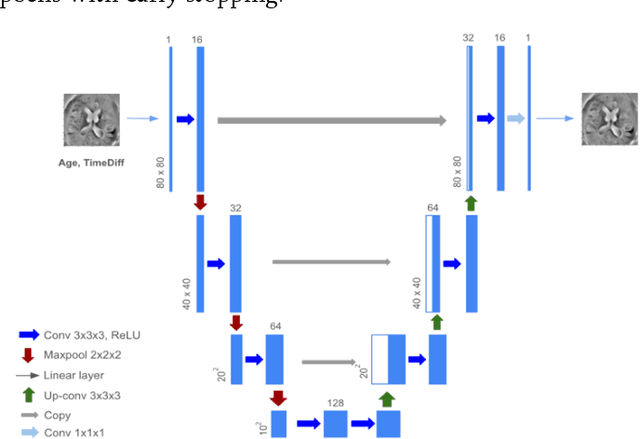
Abstract:Neurodegeneration as measured through magnetic resonance imaging (MRI) is recognized as a potential biomarker for diagnosing Alzheimer's disease (AD), but is generally considered less specific than amyloid or tau based biomarkers. Due to a large amount of variability in brain anatomy between different individuals, we hypothesize that leveraging MRI time series can help improve specificity, by treating each patient as their own baseline. Here we turn to conditional variational autoencoders to generate individualized MRI predictions given the subject's age, disease status and one previous scan. Using serial imaging data from the Alzheimer's Disease Neuroimaging Initiative, we train a novel architecture to build a latent space distribution which can be sampled from to generate future predictions of changing anatomy. This enables us to extrapolate beyond the dataset and predict MRIs up to 10 years. We evaluated the model on a held-out set from ADNI and an independent dataset (from Open Access Series of Imaging Studies). By comparing to several alternatives, we show that our model produces more individualized images with higher resolution. Further, if an individual already has a follow-up MRI, we demonstrate a usage of our model to compute a likelihood ratio classifier for disease status. In practice, the model may be able to assist in early diagnosis of AD and provide a counterfactual baseline trajectory for treatment effect estimation. Furthermore, it generates a synthetic dataset that can potentially be used for downstream tasks such as anomaly detection and classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge