Ron van Bree
Wageningen University and Research, the Netherlands
Hybrid Phenology Modeling for Predicting Temperature Effects on Tree Dormancy
Jan 28, 2025Abstract:Biophysical models offer valuable insights into climate-phenology relationships in both natural and agricultural settings. However, there are substantial structural discrepancies across models which require site-specific recalibration, often yielding inconsistent predictions under similar climate scenarios. Machine learning methods offer data-driven solutions, but often lack interpretability and alignment with existing knowledge. We present a phenology model describing dormancy in fruit trees, integrating conventional biophysical models with a neural network to address their structural disparities. We evaluate our hybrid model in an extensive case study predicting cherry tree phenology in Japan, South Korea and Switzerland. Our approach consistently outperforms both traditional biophysical and machine learning models in predicting blooming dates across years. Additionally, the neural network's adaptability facilitates parameter learning for specific tree varieties, enabling robust generalization to new sites without site-specific recalibration. This hybrid model leverages both biophysical constraints and data-driven flexibility, offering a promising avenue for accurate and interpretable phenology modeling.
Integrating processed-based models and machine learning for crop yield prediction
Jul 25, 2023Abstract:Crop yield prediction typically involves the utilization of either theory-driven process-based crop growth models, which have proven to be difficult to calibrate for local conditions, or data-driven machine learning methods, which are known to require large datasets. In this work we investigate potato yield prediction using a hybrid meta-modeling approach. A crop growth model is employed to generate synthetic data for (pre)training a convolutional neural net, which is then fine-tuned with observational data. When applied in silico, our meta-modeling approach yields better predictions than a baseline comprising a purely data-driven approach. When tested on real-world data from field trials (n=303) and commercial fields (n=77), the meta-modeling approach yields competitive results with respect to the crop growth model. In the latter set, however, both models perform worse than a simple linear regression with a hand-picked feature set and dedicated preprocessing designed by domain experts. Our findings indicate the potential of meta-modeling for accurate crop yield prediction; however, further advancements and validation using extensive real-world datasets is recommended to solidify its practical effectiveness.
Neural Prototype Trees for Interpretable Fine-grained Image Recognition
Dec 03, 2020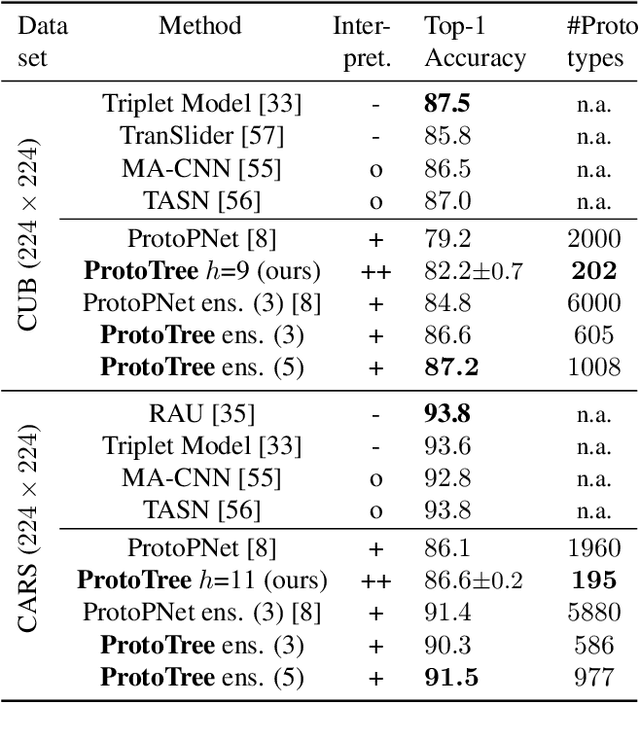

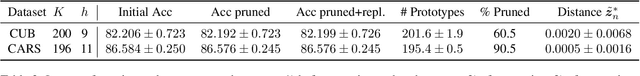

Abstract:Interpretable machine learning addresses the black-box nature of deep neural networks. Visual prototypes have been suggested for intrinsically interpretable image recognition, instead of generating post-hoc explanations that approximate a trained model. However, a large number of prototypes can be overwhelming. To reduce explanation size and improve interpretability, we propose the Neural Prototype Tree (ProtoTree), a deep learning method that includes prototypes in an interpretable decision tree to faithfully visualize the entire model. In addition to global interpretability, a path in the tree explains a single prediction. Each node in our binary tree contains a trainable prototypical part. The presence or absence of this prototype in an image determines the routing through a node. Decision making is therefore similar to human reasoning: Does the bird have a red throat? And an elongated beak? Then it's a hummingbird! We tune the accuracy-interpretability trade-off using ensembling and pruning. We apply pruning without sacrificing accuracy, resulting in a small tree with only 8 prototypes along a path to classify a bird from 200 species. An ensemble of 5 ProtoTrees achieves competitive accuracy on the CUB-200-2011 and Stanford Cars data sets. Code is available at https://github.com/M-Nauta/ProtoTree
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge