Roberto C. Sotero
Towards improving Alzheimer's intervention: a machine learning approach for biomarker detection through combining MEG and MRI pipelines
Aug 09, 2024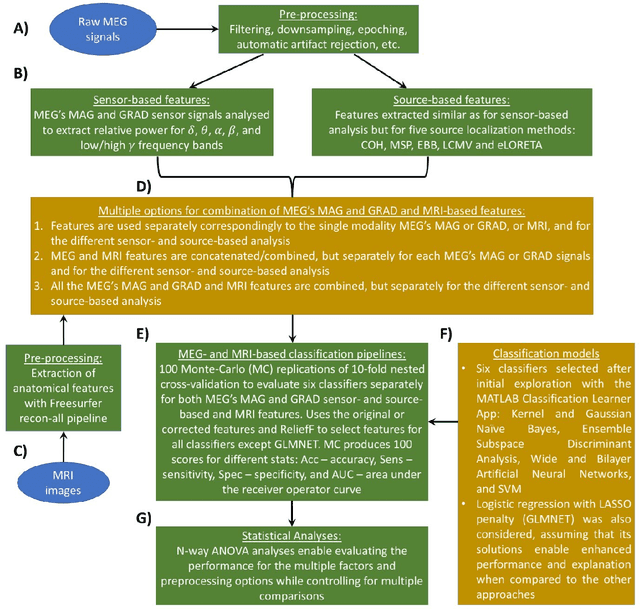
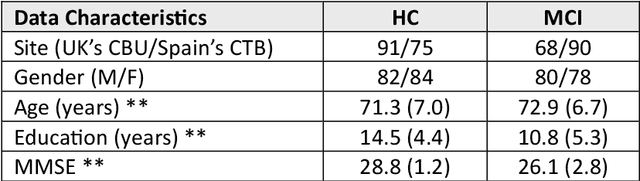

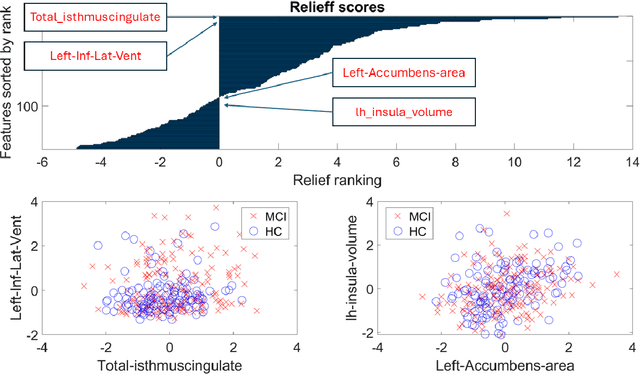
Abstract:MEG are non invasive neuroimaging techniques with excellent temporal and spatial resolution, crucial for studying brain function in dementia and Alzheimer Disease. They identify changes in brain activity at various Alzheimer stages, including preclinical and prodromal phases. MEG may detect pathological changes before clinical symptoms, offering potential biomarkers for intervention. This study evaluates classification techniques using MEG features to distinguish between healthy controls and mild cognitive impairment participants from the BioFIND study. We compare MEG based biomarkers with MRI based anatomical features, both independently and combined. We used 3 Tesla MRI and MEG data from 324 BioFIND participants;158 MCI and 166 HC. Analyses were performed using MATLAB with SPM12 and OSL toolboxes. Machine learning analyses, including 100 Monte Carlo replications of 10 fold cross validation, were conducted on sensor and source spaces. Combining MRI with MEG features achieved the best performance; 0.76 accuracy and AUC of 0.82 for GLMNET using LCMV source based MEG. MEG only analyses using LCMV and eLORETA also performed well, suggesting that combining uncorrected MEG with z-score-corrected MRI features is optimal.
Solving large-scale MEG/EEG source localization and functional connectivity problems simultaneously using state-space models
Aug 26, 2022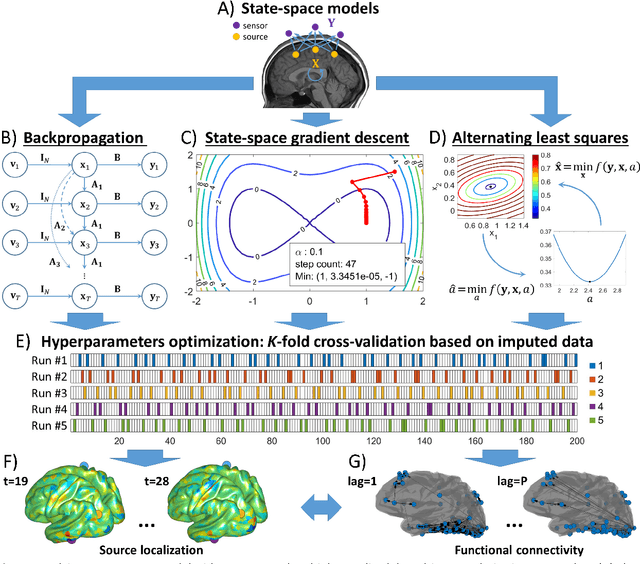
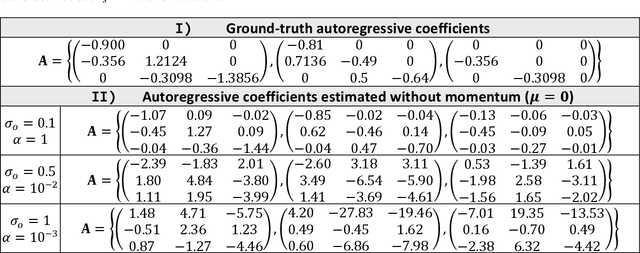
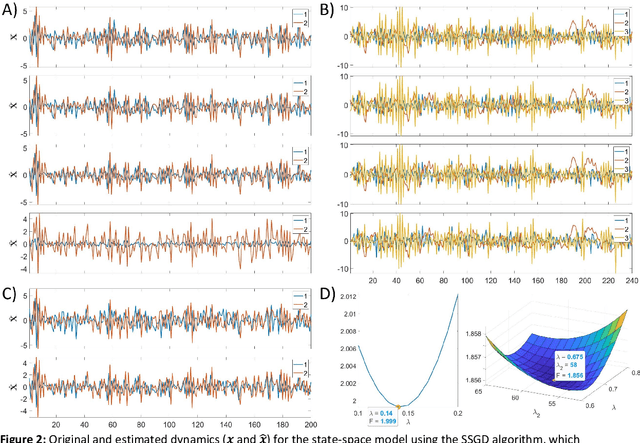
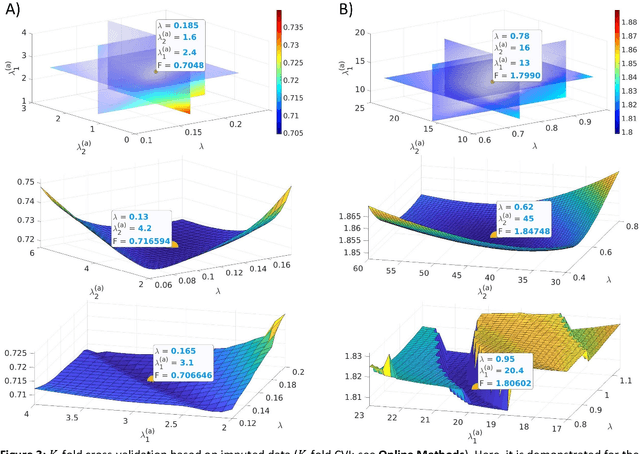
Abstract:State-space models are used in many fields when dynamics are unobserved. Popular methods such as the Kalman filter and expectation maximization enable estimation of these models but pay a high computational cost in large-scale analysis. In these approaches, sparse inverse covariance estimators can reduce the cost; however, a trade-off between enforced sparsity and increased estimation bias occurs, which demands careful consideration in low signal-to-noise ratio scenarios. We overcome these limitations by 1) Introducing multiple penalized state-space models based on data-driven regularization; 2) Implementing novel algorithms such as backpropagation, state-space gradient descent, and alternating least squares; 3) Proposing an extension of K-fold cross-validation to evaluate the regularization parameters. Finally, we solve the simultaneous brain source localization and functional connectivity problems for simulated and real MEG/EEG signals for thousands of sources on the cortical surface, demonstrating a substantial improvement over state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge