Robert Bakewell
Learning to detect chest radiographs containing lung nodules using visual attention networks
May 17, 2018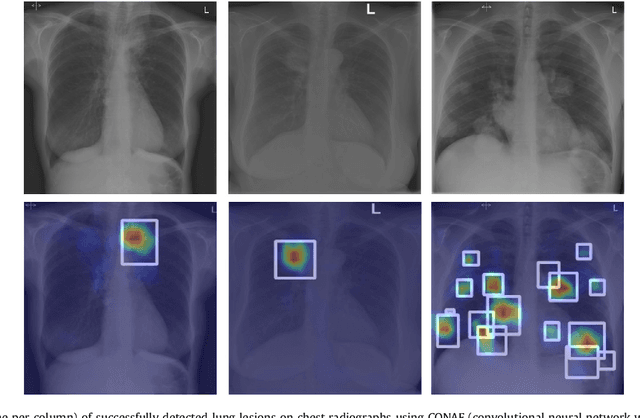
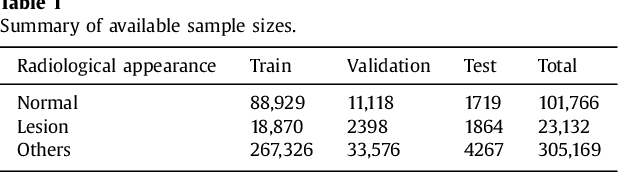
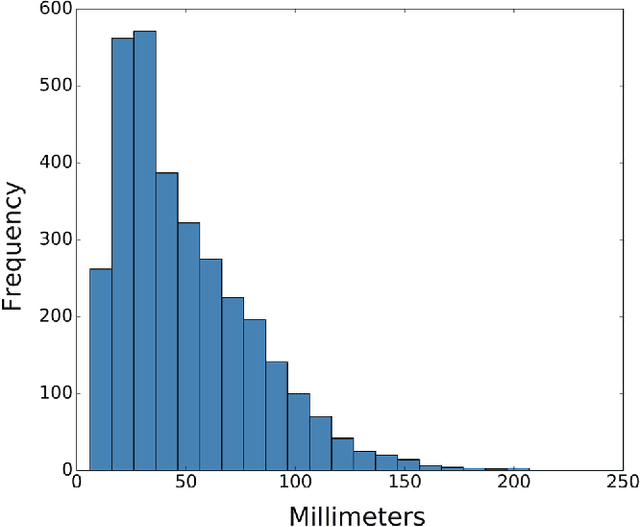

Abstract:Machine learning approaches hold great potential for the automated detection of lung nodules in chest radiographs, but training the algorithms requires vary large amounts of manually annotated images, which are difficult to obtain. Weak labels indicating whether a radiograph is likely to contain pulmonary nodules are typically easier to obtain at scale by parsing historical free-text radiological reports associated to the radiographs. Using a repositotory of over 700,000 chest radiographs, in this study we demonstrate that promising nodule detection performance can be achieved using weak labels through convolutional neural networks for radiograph classification. We propose two network architectures for the classification of images likely to contain pulmonary nodules using both weak labels and manually-delineated bounding boxes, when these are available. Annotated nodules are used at training time to deliver a visual attention mechanism informing the model about its localisation performance. The first architecture extracts saliency maps from high-level convolutional layers and compares the estimated position of a nodule against the ground truth, when this is available. A corresponding localisation error is then back-propagated along with the softmax classification error. The second approach consists of a recurrent attention model that learns to observe a short sequence of smaller image portions through reinforcement learning. When a nodule annotation is available at training time, the reward function is modified accordingly so that exploring portions of the radiographs away from a nodule incurs a larger penalty. Our empirical results demonstrate the potential advantages of these architectures in comparison to competing methodologies.
Modelling Radiological Language with Bidirectional Long Short-Term Memory Networks
Sep 27, 2016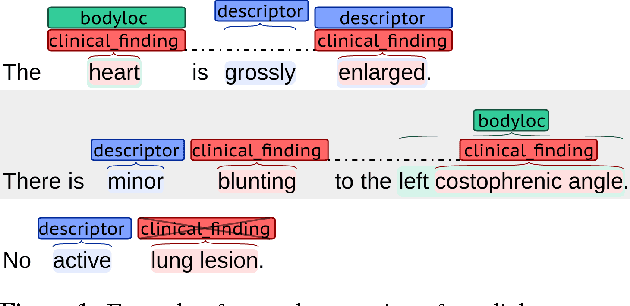
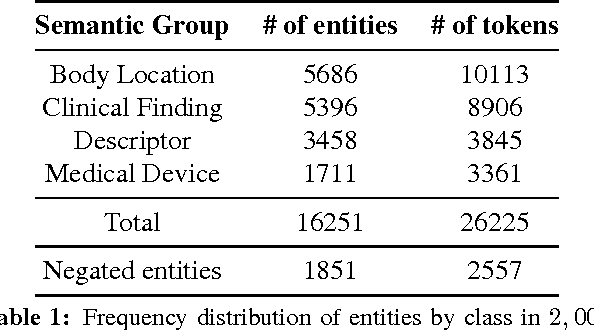
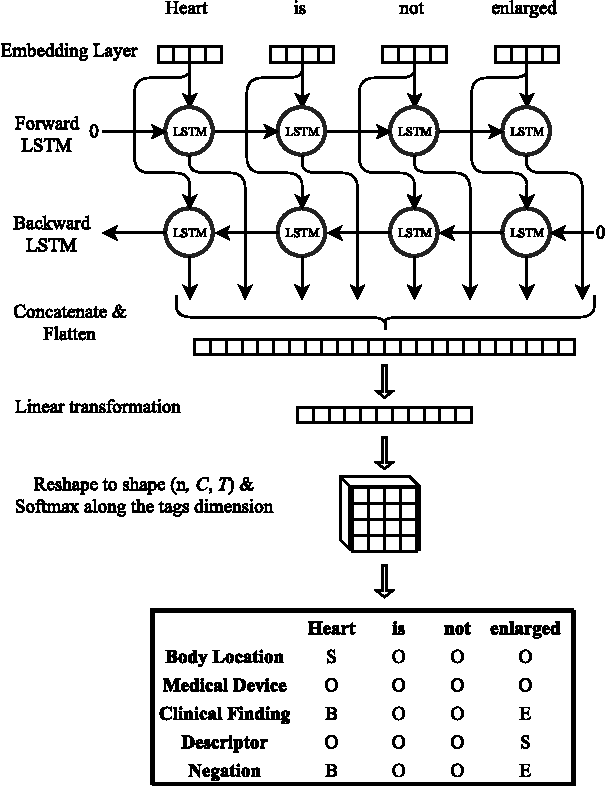
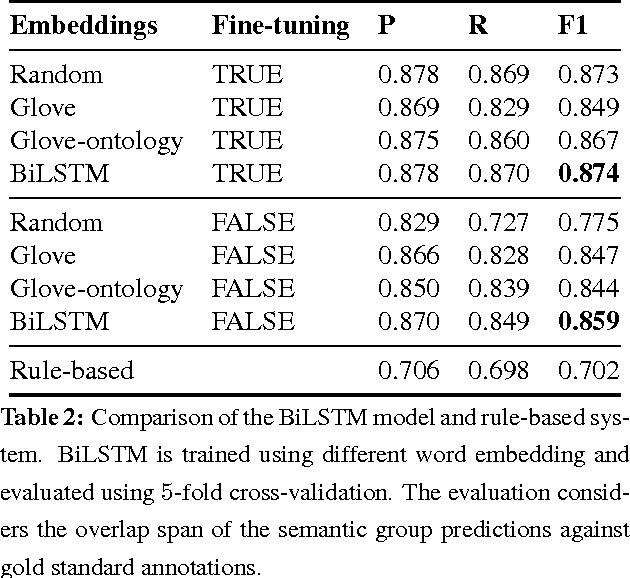
Abstract:Motivated by the need to automate medical information extraction from free-text radiological reports, we present a bi-directional long short-term memory (BiLSTM) neural network architecture for modelling radiological language. The model has been used to address two NLP tasks: medical named-entity recognition (NER) and negation detection. We investigate whether learning several types of word embeddings improves BiLSTM's performance on those tasks. Using a large dataset of chest x-ray reports, we compare the proposed model to a baseline dictionary-based NER system and a negation detection system that leverages the hand-crafted rules of the NegEx algorithm and the grammatical relations obtained from the Stanford Dependency Parser. Compared to these more traditional rule-based systems, we argue that BiLSTM offers a strong alternative for both our tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge