Ramy A. Zeineldin
Ensemble Learning and 3D Pix2Pix for Comprehensive Brain Tumor Analysis in Multimodal MRI
Dec 16, 2024



Abstract:Motivated by the need for advanced solutions in the segmentation and inpainting of glioma-affected brain regions in multi-modal magnetic resonance imaging (MRI), this study presents an integrated approach leveraging the strengths of ensemble learning with hybrid transformer models and convolutional neural networks (CNNs), alongside the innovative application of 3D Pix2Pix Generative Adversarial Network (GAN). Our methodology combines robust tumor segmentation capabilities, utilizing axial attention and transformer encoders for enhanced spatial relationship modeling, with the ability to synthesize biologically plausible brain tissue through 3D Pix2Pix GAN. This integrated approach addresses the BraTS 2023 cluster challenges by offering precise segmentation and realistic inpainting, tailored for diverse tumor types and sub-regions. The results demonstrate outstanding performance, evidenced by quantitative evaluations such as the Dice Similarity Coefficient (DSC), Hausdorff Distance (HD95) for segmentation, and Structural Similarity Index Measure (SSIM), Peak Signal-to-Noise Ratio (PSNR), and Mean-Square Error (MSE) for inpainting. Qualitative assessments further validate the high-quality, clinically relevant outputs. In conclusion, this study underscores the potential of combining advanced machine learning techniques for comprehensive brain tumor analysis, promising significant advancements in clinical decision-making and patient care within the realm of medical imaging.
Unified HT-CNNs Architecture: Transfer Learning for Segmenting Diverse Brain Tumors in MRI from Gliomas to Pediatric Tumors
Dec 11, 2024



Abstract:Accurate segmentation of brain tumors from 3D multimodal MRI is vital for diagnosis and treatment planning across diverse brain tumors. This paper addresses the challenges posed by the BraTS 2023, presenting a unified transfer learning approach that applies to a broader spectrum of brain tumors. We introduce HT-CNNs, an ensemble of Hybrid Transformers and Convolutional Neural Networks optimized through transfer learning for varied brain tumor segmentation. This method captures spatial and contextual details from MRI data, fine-tuned on diverse datasets representing common tumor types. Through transfer learning, HT-CNNs utilize the learned representations from one task to improve generalization in another, harnessing the power of pre-trained models on large datasets and fine-tuning them on specific tumor types. We preprocess diverse datasets from multiple international distributions, ensuring representativeness for the most common brain tumors. Our rigorous evaluation employs standardized quantitative metrics across all tumor types, ensuring robustness and generalizability. The proposed ensemble model achieves superior segmentation results across the BraTS validation datasets over the previous winning methods. Comprehensive quantitative evaluations using the DSC and HD95 demonstrate the effectiveness of our approach. Qualitative segmentation predictions further validate the high-quality outputs produced by our model. Our findings underscore the potential of transfer learning and ensemble approaches in medical image segmentation, indicating a substantial enhancement in clinical decision-making and patient care. Despite facing challenges related to post-processing and domain gaps, our study sets a new precedent for future research for brain tumor segmentation. The docker image for the code and models has been made publicly available, https://hub.docker.com/r/razeineldin/ht-cnns.
Multimodal CNN Networks for Brain Tumor Segmentation in MRI: A BraTS 2022 Challenge Solution
Dec 19, 2022



Abstract:Automatic segmentation is essential for the brain tumor diagnosis, disease prognosis, and follow-up therapy of patients with gliomas. Still, accurate detection of gliomas and their sub-regions in multimodal MRI is very challenging due to the variety of scanners and imaging protocols. Over the last years, the BraTS Challenge has provided a large number of multi-institutional MRI scans as a benchmark for glioma segmentation algorithms. This paper describes our contribution to the BraTS 2022 Continuous Evaluation challenge. We propose a new ensemble of multiple deep learning frameworks namely, DeepSeg, nnU-Net, and DeepSCAN for automatic glioma boundaries detection in pre-operative MRI. It is worth noting that our ensemble models took first place in the final evaluation on the BraTS testing dataset with Dice scores of 0.9294, 0.8788, and 0.8803, and Hausdorf distance of 5.23, 13.54, and 12.05, for the whole tumor, tumor core, and enhancing tumor, respectively. Furthermore, the proposed ensemble method ranked first in the final ranking on another unseen test dataset, namely Sub-Saharan Africa dataset, achieving mean Dice scores of 0.9737, 0.9593, and 0.9022, and HD95 of 2.66, 1.72, 3.32 for the whole tumor, tumor core, and enhancing tumor, respectively. The docker image for the winning submission is publicly available at (https://hub.docker.com/r/razeineldin/camed22).
Self-supervised iRegNet for the Registration of Longitudinal Brain MRI of Diffuse Glioma Patients
Nov 20, 2022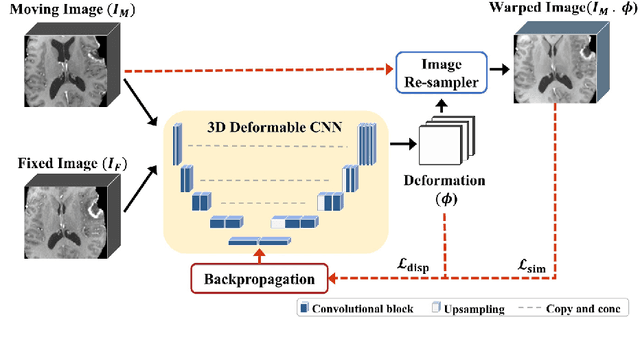
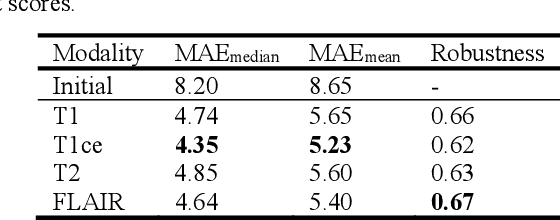
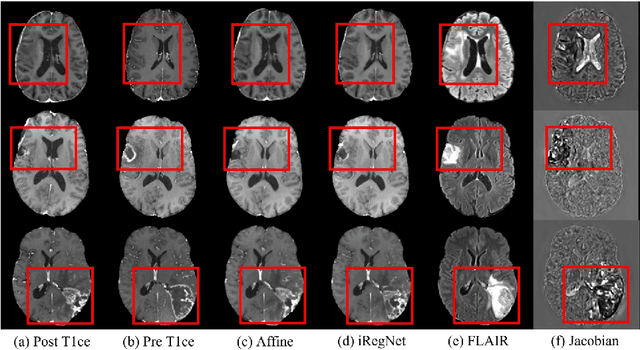
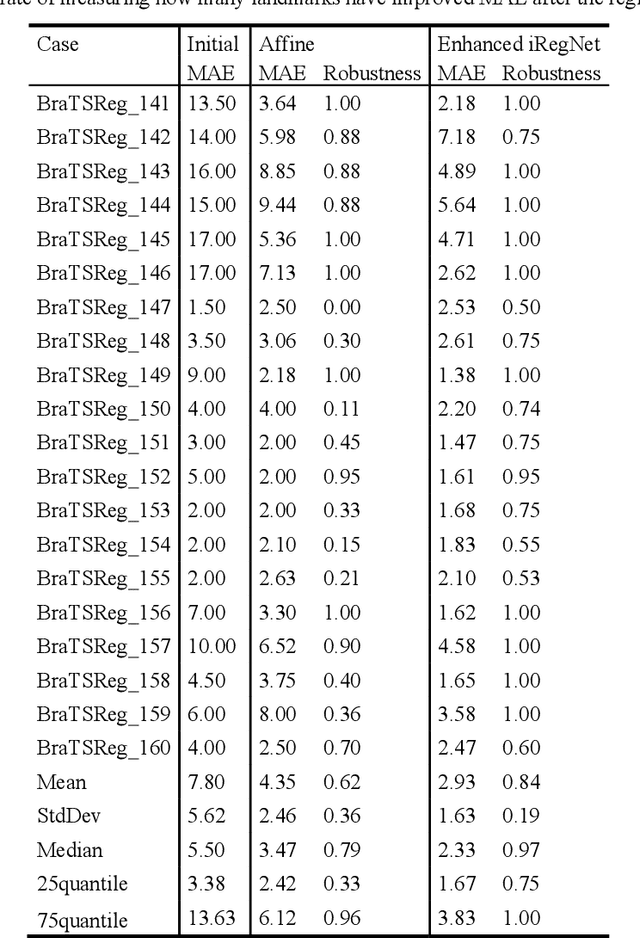
Abstract:Reliable and accurate registration of patient-specific brain magnetic resonance imaging (MRI) scans containing pathologies is challenging due to tissue appearance changes. This paper describes our contribution to the Registration of the longitudinal brain MRI task of the Brain Tumor Sequence Registration Challenge 2022 (BraTS-Reg 2022). We developed an enhanced unsupervised learning-based method that extends the iRegNet. In particular, incorporating an unsupervised learning-based paradigm as well as several minor modifications to the network pipeline, allows the enhanced iRegNet method to achieve respectable results. Experimental findings show that the enhanced self-supervised model is able to improve the initial mean median registration absolute error (MAE) from 8.20 (7.62) mm to the lowest value of 3.51 (3.50) for the training set while achieving an MAE of 2.93 (1.63) mm for the validation set. Additional qualitative validation of this study was conducted through overlaying pre-post MRI pairs before and after the de-formable registration. The proposed method scored 5th place during the testing phase of the MICCAI BraTS-Reg 2022 challenge. The docker image to reproduce our BraTS-Reg submission results will be publicly available.
Ensemble CNN Networks for GBM Tumors Segmentation using Multi-parametric MRI
Dec 27, 2021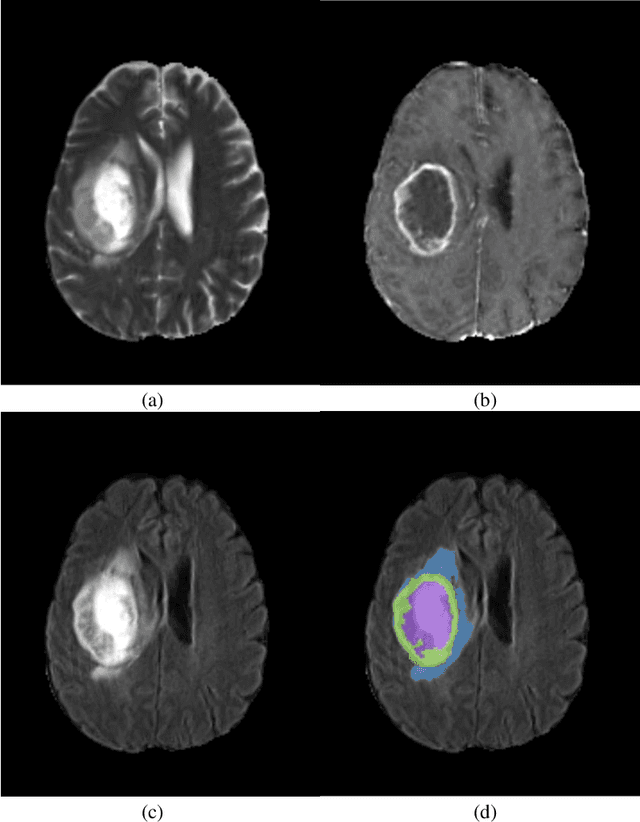
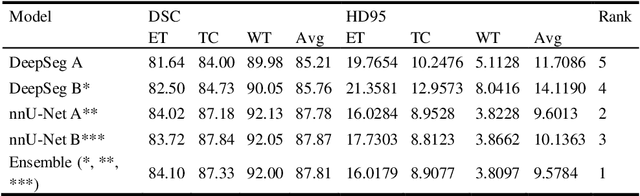
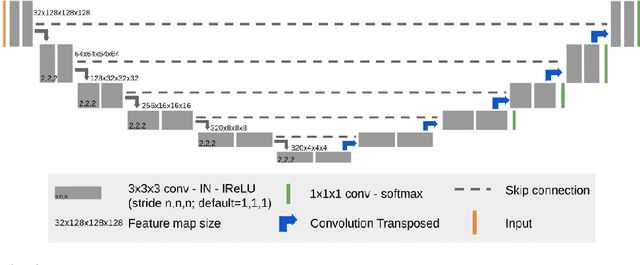
Abstract:Glioblastomas are the most aggressive fast-growing primary brain cancer which originate in the glial cells of the brain. Accurate identification of the malignant brain tumor and its sub-regions is still one of the most challenging problems in medical image segmentation. The Brain Tumor Segmentation Challenge (BraTS) has been a popular benchmark for automatic brain glioblastomas segmentation algorithms since its initiation. In this year, BraTS 2021 challenge provides the largest multi-parametric (mpMRI) dataset of 2,000 pre-operative patients. In this paper, we propose a new aggregation of two deep learning frameworks namely, DeepSeg and nnU-Net for automatic glioblastoma recognition in pre-operative mpMRI. Our ensemble method obtains Dice similarity scores of 92.00, 87.33, and 84.10 and Hausdorff Distances of 3.81, 8.91, and 16.02 for the enhancing tumor, tumor core, and whole tumor regions, respectively, on the BraTS 2021 validation set, ranking us among the top ten teams. These experimental findings provide evidence that it can be readily applied clinically and thereby aiding in the brain cancer prognosis, therapy planning, and therapy response monitoring. A docker image for reproducing our segmentation results is available online at (https://hub.docker.com/r/razeineldin/deepseg21).
DeepSeg: Deep Neural Network Framework for Automatic Brain Tumor Segmentation using Magnetic Resonance FLAIR Images
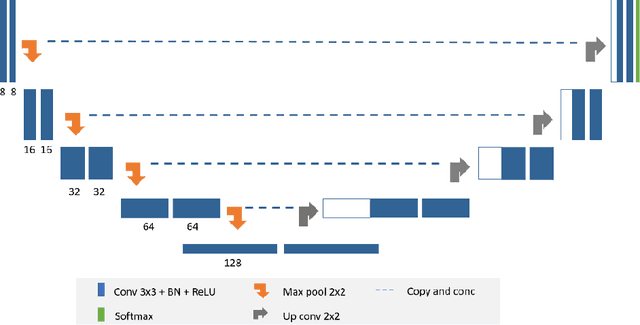
Apr 26, 2020Abstract:Purpose: Gliomas are the most common and aggressive type of brain tumors due to their infiltrative nature and rapid progression. The process of distinguishing tumor boundaries from healthy cells is still a challenging task in the clinical routine. Fluid-Attenuated Inversion Recovery (FLAIR) MRI modality can provide the physician with information about tumor infiltration. Therefore, this paper proposes a new generic deep learning architecture; namely DeepSeg for fully automated detection and segmentation of the brain lesion using FLAIR MRI data. Methods: The developed DeepSeg is a modular decoupling framework. It consists of two connected core parts based on an encoding and decoding relationship. The encoder part is a convolutional neural network (CNN) responsible for spatial information extraction. The resulting semantic map is inserted into the decoder part to get the full resolution probability map. Based on modified U-Net architecture, different CNN models such as Residual Neural Network (ResNet), Dense Convolutional Network (DenseNet), and NASNet have been utilized in this study. Results: The proposed deep learning architectures have been successfully tested and evaluated on-line based on MRI datasets of Brain Tumor Segmentation (BraTS 2019) challenge, including s336 cases as training data and 125 cases for validation data. The dice and Hausdorff distance scores of obtained segmentation results are about 0.81 to 0.84 and 9.8 to 19.7 correspondingly. Conclusion: This study showed successful feasibility and comparative performance of applying different deep learning models in a new DeepSeg framework for automated brain tumor segmentation in FLAIR MR images. The proposed DeepSeg is open-source and freely available at https://github.com/razeineldin/DeepSeg/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge