Ramakrishnan Mukundan
University of Canterbury
Grey Level Texture Features for Segmentation of Chromogenic Dye RNAscope From Breast Cancer Tissue
Jan 29, 2024Abstract:Chromogenic RNAscope dye and haematoxylin staining of cancer tissue facilitates diagnosis of the cancer type and subsequent treatment, and fits well into existing pathology workflows. However, manual quantification of the RNAscope transcripts (dots), which signify gene expression, is prohibitively time consuming. In addition, there is a lack of verified supporting methods for quantification and analysis. This paper investigates the usefulness of gray level texture features for automatically segmenting and classifying the positions of RNAscope transcripts from breast cancer tissue. Feature analysis showed that a small set of gray level features, including Gray Level Dependence Matrix and Neighbouring Gray Tone Difference Matrix features, were well suited for the task. The automated method performed similarly to expert annotators at identifying the positions of RNAscope transcripts, with an F1-score of 0.571 compared to the expert inter-rater F1-score of 0.596. These results demonstrate the potential of gray level texture features for automated quantification of RNAscope in the pathology workflow.
A Framework for Visually Realistic Multi-robot Simulation in Natural Environment
Aug 06, 2017



Abstract:This paper presents a generalized framework for the simulation of multiple robots and drones in highly realistic models of natural environments. The proposed simulation architecture uses the Unreal Engine4 for generating both optical and depth sensor outputs from any position and orientation within the environment and provides several key domain specific simulation capabilities. Various components and functionalities of the system have been discussed in detail. The simulation engine also allows users to test and validate a wide range of computer vision algorithms involving different drone configurations under many types of environmental effects such as wind gusts. The paper demonstrates the effectiveness of the system by giving experimental results for a test scenario where one drone tracks the simulated motion of another in a complex natural environment.
Her2 Challenge Contest: A Detailed Assessment of Automated Her2 Scoring Algorithms in Whole Slide Images of Breast Cancer Tissues
Jul 24, 2017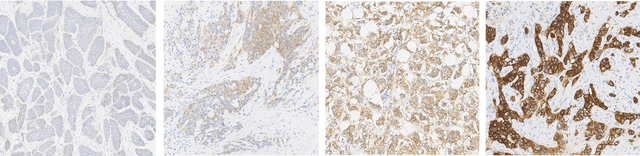
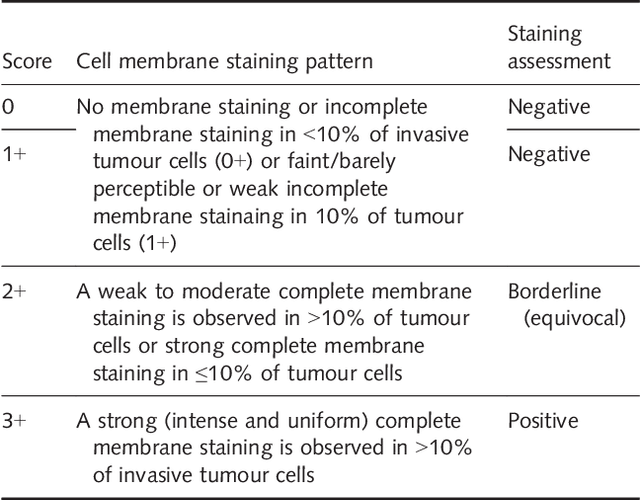
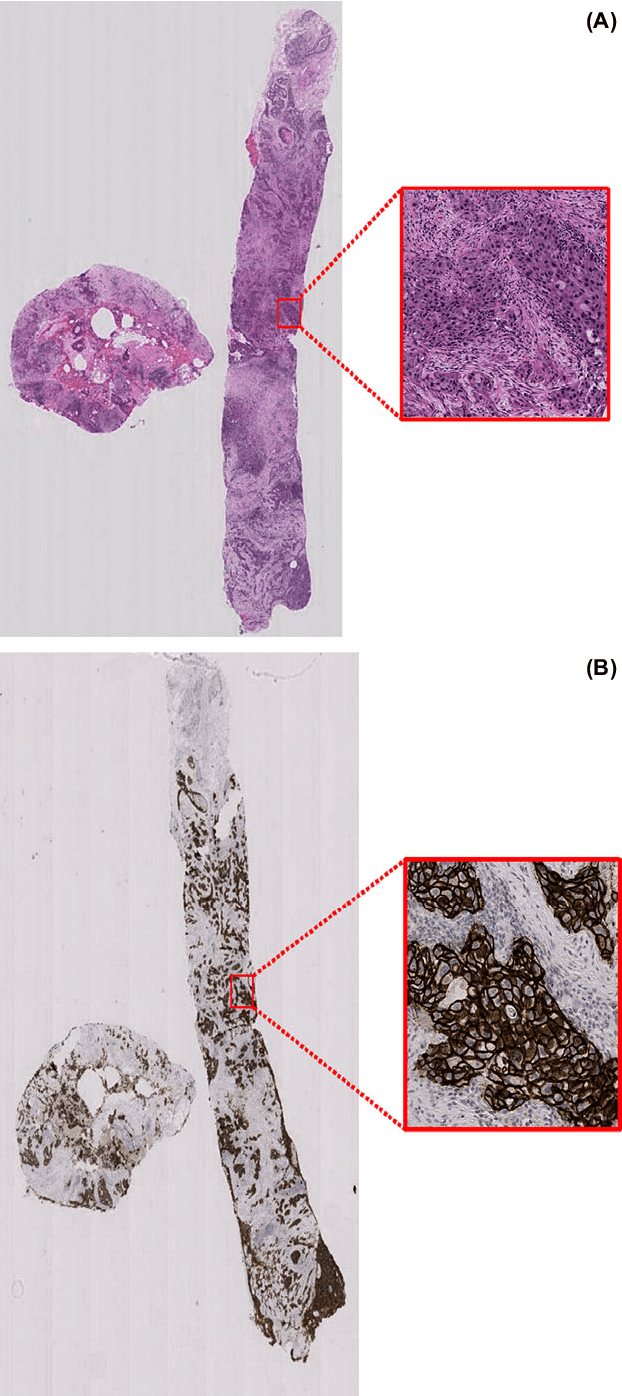
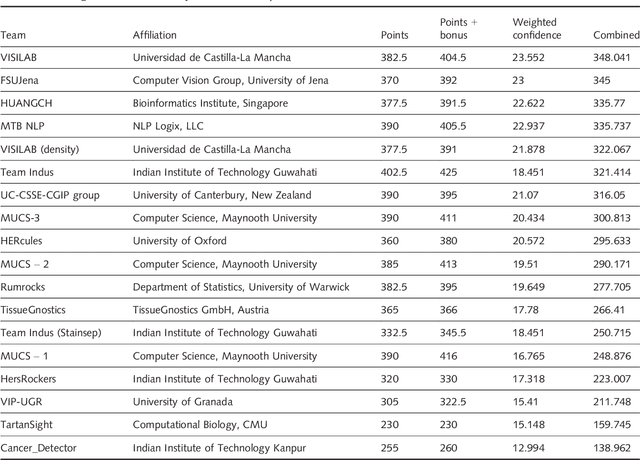
Abstract:Evaluating expression of the Human epidermal growth factor receptor 2 (Her2) by visual examination of immunohistochemistry (IHC) on invasive breast cancer (BCa) is a key part of the diagnostic assessment of BCa due to its recognised importance as a predictive and prognostic marker in clinical practice. However, visual scoring of Her2 is subjective and consequently prone to inter-observer variability. Given the prognostic and therapeutic implications of Her2 scoring, a more objective method is required. In this paper, we report on a recent automated Her2 scoring contest, held in conjunction with the annual PathSoc meeting held in Nottingham in June 2016, aimed at systematically comparing and advancing the state-of-the-art Artificial Intelligence (AI) based automated methods for Her2 scoring. The contest dataset comprised of digitised whole slide images (WSI) of sections from 86 cases of invasive breast carcinoma stained with both Haematoxylin & Eosin (H&E) and IHC for Her2. The contesting algorithms automatically predicted scores of the IHC slides for an unseen subset of the dataset and the predicted scores were compared with the 'ground truth' (a consensus score from at least two experts). We also report on a simple Man vs Machine contest for the scoring of Her2 and show that the automated methods could beat the pathology experts on this contest dataset. This paper presents a benchmark for comparing the performance of automated algorithms for scoring of Her2. It also demonstrates the enormous potential of automated algorithms in assisting the pathologist with objective IHC scoring.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge