Ram J. Zaveri
Few-shot adaptation for morphology-independent cell instance segmentation
Feb 27, 2024



Abstract:Microscopy data collections are becoming larger and more frequent. Accurate and precise quantitative analysis tools like cell instance segmentation are necessary to benefit from them. This is challenging due to the variability in the data, which requires retraining the segmentation model to maintain high accuracy on new collections. This is needed especially for segmenting cells with elongated and non-convex morphology like bacteria. We propose to reduce the amount of annotation and computing power needed for retraining the model by introducing a few-shot domain adaptation approach that requires annotating only one to five cells of the new data to process and that quickly adapts the model to maintain high accuracy. Our results show a significant boost in accuracy after adaptation to very challenging bacteria datasets.
Fine-Grained Visual Classification of Plant Species In The Wild: Object Detection as A Reinforced Means of Attention
Jun 03, 2021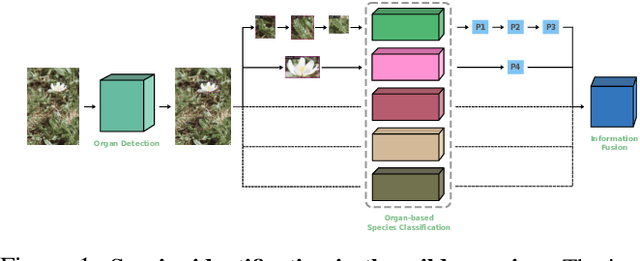
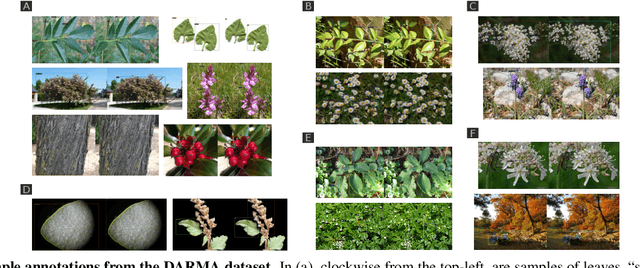

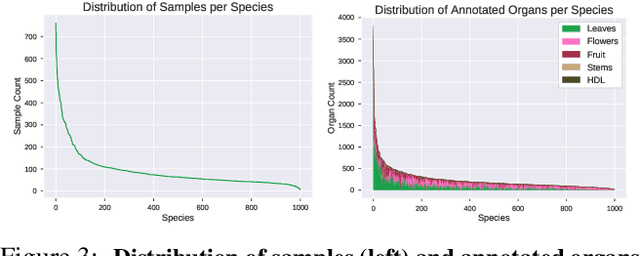
Abstract:Plant species identification in the wild is a difficult problem in part due to the high variability of the input data, but also because of complications induced by the long-tail effects of the datasets distribution. Inspired by the most recent fine-grained visual classification approaches which are based on attention to mitigate the effects of data variability, we explore the idea of using object detection as a form of attention. We introduce a bottom-up approach based on detecting plant organs and fusing the predictions of a variable number of organ-based species classifiers. We also curate a new dataset with a long-tail distribution for evaluating plant organ detection and organ-based species identification, which is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge