Rahul Desai
The Many Faces of Robustness: A Critical Analysis of Out-of-Distribution Generalization
Jun 29, 2020
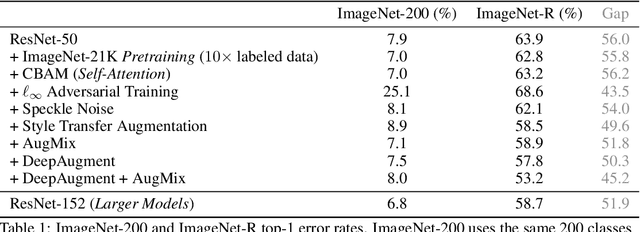
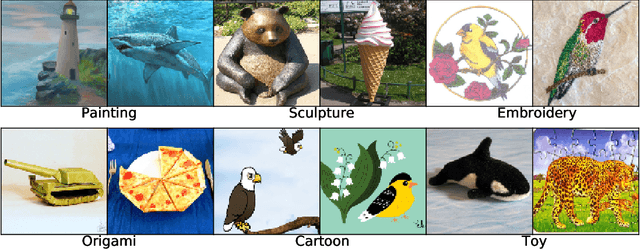
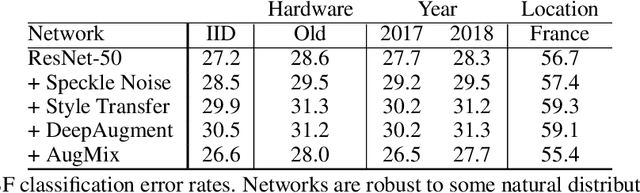
Abstract:We introduce three new robustness benchmarks consisting of naturally occurring distribution changes in image style, geographic location, camera operation, and more. Using our benchmarks, we take stock of previously proposed hypotheses for out-of-distribution robustness and put them to the test. We find that using larger models and synthetic data augmentation can improve robustness on real-world distribution shifts, contrary to claims in prior work. Motivated by this, we introduce a new data augmentation method which advances the state-of-the-art and outperforms models pretrained with 1000x more labeled data. We find that some methods consistently help with distribution shifts in texture and local image statistics, but these methods do not help with some other distribution shifts like geographic changes. We conclude that future research must study multiple distribution shifts simultaneously.
NER Models Using Pre-training and Transfer Learning for Healthcare
Oct 23, 2019



Abstract:In this paper, we present our approach to extract structured information from unstructured Electronic Health Records (EHR) [2] to study adverse drug reactions on patients, due to chemicals in their products. Our solution uses a combination of Natural Language Processing (NLP) techniques and a web-based annotation tool to optimize the performance of a custom Named Entity Recognition (NER) [1] model trained on a limited amount of EHR training data. We showcase a combination of tools and techniques leveraging the recent advancements in NLP aimed at targeting domain shifts by applying transfer learning and language model pre-training techniques [3]. We present a comparison of our technique to the base models available and show the effective increase in performance of the NER model and the reduction in time to annotate data. A key observation of the results presented is that the F1 score of model (0.734) trained with our approach with just 50% of available training data outperforms the F1 score of the blank spaCy model (0.704) trained with 100% of the available training data. We also demonstrate an annotation tool to minimize domain expert time and the manual effort required to generate such a training dataset. Further, we plan to release the annotated dataset as well as the pre-trained model to the community to further research in medical health records.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge